 |
Reference documentation for deal.II version 9.2.0
|
 |
Reference documentation for deal.II version 9.2.0
|
This tutorial depends on step-10.
| Table of contents | |
|---|---|
The problem we will be considering is the solution of Laplace's problem with Neumann boundary conditions only:
\begin{eqnarray*} -\Delta u &=& f \qquad \mathrm{in}\ \Omega, \\ \partial_n u &=& g \qquad \mathrm{on}\ \partial\Omega. \end{eqnarray*}
It is well known that if this problem is to have a solution, then the forces need to satisfy the compatibility condition
\[ \int_\Omega f\; dx + \int_{\partial\Omega} g\; ds = 0. \]
We will consider the special case that \(\Omega\) is the circle of radius 1 around the origin, and \(f=-2\), \(g=1\). This choice satisfies the compatibility condition.
The compatibility condition allows a solution of the above equation, but it nevertheless retains an ambiguity: since only derivatives of the solution appear in the equations, the solution is only determined up to a constant. For this reason, we have to pose another condition for the numerical solution, which fixes this constant.
For this, there are various possibilities:
Fix one node of the discretization to zero or any other fixed value. This amounts to an additional condition \(u_h(x_0)=0\). Although this is common practice, it is not necessarily a good idea, since we know that the solutions of Laplace's equation are only in \(H^1\), which does not allow for the definition of point values because it is not a subset of the continuous functions. Therefore, even though fixing one node is allowed for discretized functions, it is not for continuous functions, and one can often see this in a resulting error spike at this point in the numerical solution.
Fixing the mean value over the domain to zero or any other value. This is allowed on the continuous level, since \(H^1(\Omega)\subset L^1(\Omega)\) by Sobolev's inequality, and thus also on the discrete level since we there only consider subsets of \(H^1\).
We will choose the last possibility, since we want to demonstrate another technique with it.
While this describes the problem to be solved, we still have to figure out how to implement it. Basically, except for the additional mean value constraint, we have solved this problem several times, using Dirichlet boundary values, and we only need to drop the treatment of Dirichlet boundary nodes. The use of higher order mappings is also rather trivial and will be explained at the various places where we use it; in almost all conceivable cases, you will only consider the objects describing mappings as a black box which you need not worry about, because their only uses seem to be to be passed to places deep inside the library where functions know how to handle them (i.e. in the FEValues classes and their descendants).
The tricky point in this program is the use of the mean value constraint. Fortunately, there is a class in the library which knows how to handle such constraints, and we have used it quite often already, without mentioning its generality. Note that if we assume that the boundary nodes are spaced equally along the boundary, then the mean value constraint
\[ \int_{\partial \Omega} u(x) \; ds = 0 \]
can be written as
\[ \sum_{i\in\partial\Omega_h} u_i = 0, \]
where the sum shall run over all degree of freedom indices which are located on the boundary of the computational domain. Let us denote by \(i_0\) that index on the boundary with the lowest number (or any other conveniently chosen index), then the constraint can also be represented by
\[ u_{i_0} = \sum_{i\in\partial\Omega_h\backslash i_0} -u_i. \]
This, luckily, is exactly the form of constraints for which the AffineConstraints class was designed. Note that we have used this class in several previous examples for the representation of hanging nodes constraints, which also have this form: there, the middle vertex shall have the mean of the values of the adjacent vertices. In general, the AffineConstraints class is designed to handle affine constraints of the form
\[ CU = b \]
where \(C\) denotes a matrix, \(b\) denotes a vector, and \(U\) the vector of nodal values. In this case, since \(C\) represents one homogeneous constraint, \(b\) is the zero vector.
In this example, the mean value along the boundary allows just such a representation, with \(C\) being a matrix with just one row (i.e. there is only one constraint). In the implementation, we will create an AffineConstraints object, add one constraint (i.e. add another row to the matrix) referring to the first boundary node \(i_0\), and insert the weights with which all the other nodes contribute, which in this example happens to be just \(-1\).
Later, we will use this object to eliminate the first boundary node from the linear system of equations, reducing it to one which has a solution without the ambiguity of the constant shift value. One of the problems of the implementation will be that the explicit elimination of this node results in a number of additional elements in the matrix, of which we do not know in advance where they are located and how many additional entries will be in each of the rows of the matrix. We will show how we can use an intermediate object to work around this problem.
But now on to the implementation of the program solving this problem...
As usual, the program starts with a rather long list of include files which you are probably already used to by now:
Just this one is new: it declares a class DynamicSparsityPattern, which we will use and explain further down below.
We will make use of the std::find algorithm of the C++ standard library, so we have to include the following file for its declaration:
The last step is as in all previous programs:
Then we declare a class which represents the solution of a Laplace problem. As this example program is based on step-5, the class looks rather the same, with the sole structural difference that the functions assemble_system now calls solve itself, and is thus called assemble_and_solve, and that the output function was dropped since the solution function is so boring that it is not worth being viewed.
The only other noteworthy change is that the constructor takes a value representing the polynomial degree of the mapping to be used later on, and that it has another member variable representing exactly this mapping. In general, this variable will occur in real applications at the same places where the finite element is declared or used.
Construct such an object, by initializing the variables. Here, we use linear finite elements (the argument to the fe variable denotes the polynomial degree), and mappings of given order. Print to screen what we are about to do.
The first task is to set up the variables for this problem. This includes generating a valid DoFHandler object, as well as the sparsity patterns for the matrix, and the object representing the constraints that the mean value of the degrees of freedom on the boundary be zero.
The first task is trivial: generate an enumeration of the degrees of freedom, and initialize solution and right hand side vector to their correct sizes:
Next task is to construct the object representing the constraint that the mean value of the degrees of freedom on the boundary shall be zero. For this, we first want a list of those nodes which are actually at the boundary. The DoFTools namespace has a function that returns an array of Boolean values where true indicates that the node is at the boundary. The second argument denotes a mask selecting which components of vector valued finite elements we want to be considered. This sort of information is encoded using the ComponentMask class (see also GlossComponentMask). Since we have a scalar finite element anyway, this mask in reality should have only one entry with a true value. However, the ComponentMask class has semantics that allow it to represents a mask of indefinite size whose every element equals true when one just default constructs such an object, so this is what we'll do here.
Now first for the generation of the constraints: as mentioned in the introduction, we constrain one of the nodes on the boundary by the values of all other DoFs on the boundary. So, let us first pick out the first boundary node from this list. We do that by searching for the first true value in the array (note that std::find returns an iterator to this element), and computing its distance to the overall first element in the array to get its index:
Then generate a constraints object with just this one constraint. First clear all previous content (which might reside there from the previous computation on a once coarser grid), then add this one line constraining the first_boundary_dof to the sum of other boundary DoFs each with weight -1. Finally, close the constraints object, i.e. do some internal bookkeeping on it for faster processing of what is to come later:
Next task is to generate a sparsity pattern. This is indeed a tricky task here. Usually, we just call DoFTools::make_sparsity_pattern and condense the result using the hanging node constraints. We have no hanging node constraints here (since we only refine globally in this example), but we have this global constraint on the boundary. This poses one severe problem in this context: the SparsityPattern class wants us to state beforehand the maximal number of entries per row, either for all rows or for each row separately. There are functions in the library which can tell you this number in case you just have hanging node constraints (namely DoFHandler::max_couplings_between_dofs), but how is this for the present case? The difficulty arises because the elimination of the constrained degree of freedom requires a number of additional entries in the matrix at places that are not so simple to determine. We would therefore have a problem had we to give a maximal number of entries per row here.
Since this can be so difficult that no reasonable answer can be given that allows allocation of only a reasonable amount of memory, there is a class DynamicSparsityPattern, that can help us out here. It does not require that we know in advance how many entries rows could have, but allows just about any length. It is thus significantly more flexible in case you do not have good estimates of row lengths, however at the price that building up such a pattern is also significantly more expensive than building up a pattern for which you had information in advance. Nevertheless, as we have no other choice here, we'll just build such an object by initializing it with the dimensions of the matrix and calling another function DoFTools::make_sparsity_pattern to get the sparsity pattern due to the differential operator, then condense it with the constraints object which adds those positions in the sparsity pattern that are required for the elimination of the constraint.
Finally, once we have the full pattern, we can initialize an object of type SparsityPattern from it and in turn initialize the matrix with it. Note that this is actually necessary, since the DynamicSparsityPattern is so inefficient compared to the SparsityPattern class due to the more flexible data structures it has to use, that we can impossibly base the sparse matrix class on it, but rather need an object of type SparsityPattern, which we generate by copying from the intermediate object.
As a further sidenote, you will notice that we do not explicitly have to compress the sparsity pattern here. This, of course, is due to the fact that the copy_from function generates a compressed object right from the start, to which you cannot add new entries anymore. The compress call is therefore implicit in the copy_from call.
The next function then assembles the linear system of equations, solves it, and evaluates the solution. This then makes three actions, and we will put them into eight true statements (excluding declaration of variables, and handling of temporary vectors). Thus, this function is something for the very lazy. Nevertheless, the functions called are rather powerful, and through them this function uses a good deal of the whole library. But let's look at each of the steps.
First, we have to assemble the matrix and the right hand side. In all previous examples, we have investigated various ways how to do this manually. However, since the Laplace matrix and simple right hand sides appear so frequently in applications, the library provides functions for actually doing this for you, i.e. they perform the loop over all cells, setting up the local matrices and vectors, and putting them together for the end result.
The following are the two most commonly used ones: creation of the Laplace matrix and creation of a right hand side vector from body or boundary forces. They take the mapping object, the DoFHandler object representing the degrees of freedom and the finite element in use, a quadrature formula to be used, and the output object. The function that creates a right hand side vector also has to take a function object describing the (continuous) right hand side function.
Let us look at the way the matrix and body forces are integrated:
That's quite simple, right?
Two remarks are in order, though: First, these functions are used in a lot of contexts. Maybe you want to create a Laplace or mass matrix for a vector values finite element; or you want to use the default Q1 mapping; or you want to assembled the matrix with a coefficient in the Laplace operator. For this reason, there are quite a large number of variants of these functions in the MatrixCreator and MatrixTools namespaces. Whenever you need a slightly different version of these functions than the ones called above, it is certainly worthwhile to take a look at the documentation and to check whether something fits your needs.
The second remark concerns the quadrature formula we use: we want to integrate over bilinear shape functions, so we know that we have to use at least an order two Gauss quadrature formula. On the other hand, we want the quadrature rule to have at least the order of the boundary approximation. Since the order of Gauss rule with \(r\) points is \(2r - 1\), and the order of the boundary approximation using polynomials of degree \(p\) is \(p+1\), we know that \(2r \geq p\). Since r has to be an integer and (as mentioned above) has to be at least \(2\), this makes up for the formula above computing gauss_degree.
Since the generation of the body force contributions to the right hand side vector was so simple, we do that all over again for the boundary forces as well: allocate a vector of the right size and call the right function. The boundary function has constant values, so we can generate an object from the library on the fly, and we use the same quadrature formula as above, but this time of lower dimension since we integrate over faces now instead of cells:
Then add the contributions from the boundary to those from the interior of the domain:
For assembling the right hand side, we had to use two different vector objects, and later add them together. The reason we had to do so is that the VectorTools::create_right_hand_side and VectorTools::create_boundary_right_hand_side functions first clear the output vector, rather than adding up their results to previous contents. This can reasonably be called a design flaw in the library made in its infancy, but unfortunately things are as they are for some time now and it is difficult to change such things that silently break existing code, so we have to live with that.
Now, the linear system is set up, so we can eliminate the one degree of freedom which we constrained to the other DoFs on the boundary for the mean value constraint from matrix and right hand side vector, and solve the system. After that, distribute the constraints again, which in this case means setting the constrained degree of freedom to its proper value
Finally, evaluate what we got as solution. As stated in the introduction, we are interested in the H1 semi-norm of the solution. Here, as well, we have a function in the library that does this, although in a slightly non-obvious way: the VectorTools::integrate_difference function integrates the norm of the difference between a finite element function and a continuous function. If we therefore want the norm of a finite element field, we just put the continuous function to zero. Note that this function, just as so many other ones in the library as well, has at least two versions, one which takes a mapping as argument (which we make us of here), and the one which we have used in previous examples which implicitly uses MappingQ1. Also note that we take a quadrature formula of one degree higher, in order to avoid superconvergence effects where the solution happens to be especially close to the exact solution at certain points (we don't know whether this might be the case here, but there are cases known of this, and we just want to make sure):
Then, the function just called returns its results as a vector of values each of which denotes the norm on one cell. To get the global norm, we do the following:
Last task – generate output:
The following function solving the linear system of equations is copied from step-5 and is explained there in some detail:
Next, we write the solution as well as the material ids to a VTU file. This is similar to what was done in many other tutorial programs. The new ingredient presented in this tutorial program is that we want to ensure that the data written to the file used for visualization is actually a faithful representation of what is used internally by deal.II. That is because most of the visualization data formats only represent cells by their vertex coordinates, but have no way of representing the curved boundaries that are used in deal.II when using higher order mappings – in other words, what you see in the visualization tool is not actually what you are computing on. (The same, incidentally, is true when using higher order shape functions: Most visualization tools only render bilinear/trilinear representations. This is discussed in detail in DataOut::build_patches().)
So we need to ensure that a high-order representation is written to the file. We need to consider two particular topics. Firstly, we tell the DataOut object via the DataOutBase::VtkFlags that we intend to interpret the subdivisions of the elements as a high-order Lagrange polynomial rather than a collection of bilinear patches. Recent visualization programs, like ParaView version 5.5 or newer, can then render a high-order solution (see a wiki page for more details). Secondly, we need to make sure that the mapping is passed to the DataOut::build_patches() method. Finally, the DataOut class only prints curved faces for boundary cells by default, so we need to ensure that also inner cells are printed in a curved representation via the mapping.
Finally the main function controlling the different steps to be performed. Its content is rather straightforward, generating a triangulation of a circle, associating a boundary to it, and then doing several cycles on subsequently finer grids. Note again that we have put mesh refinement into the loop header; this may be something for a test program, but for real applications you should consider that this implies that the mesh is refined after the loop is executed the last time since the increment clause (the last part of the three-parted loop header) is executed before the comparison part (the second one), which may be rather costly if the mesh is already quite refined. In that case, you should arrange code such that the mesh is not further refined after the last loop run (or you should do it at the beginning of each run except for the first one).
After all the data is generated, write a table of results to the screen:
Finally the main function. It's structure is the same as that used in several of the previous examples, so probably needs no more explanation.
This is the main loop, doing the computations with mappings of linear through cubic mappings. Note that since we need the object of type LaplaceProblem<2> only once, we do not even name it, but create an unnamed such object and call the run function of it, subsequent to which it is immediately destroyed again.
This is what the program outputs:
As we expected, the convergence order for each of the different mappings is clearly quadratic in the mesh size. What is interesting, though, is that the error for a bilinear mapping (i.e. degree 1) is more than three times larger than that for the higher order mappings; it is therefore clearly advantageous in this case to use a higher order mapping, not because it improves the order of convergence but just to reduce the constant before the convergence order. On the other hand, using a cubic mapping only improves the result further by insignificant amounts, except on very coarse grids.
We can also visualize the underlying meshes by using, for instance, ParaView. The image below shows initial meshes for different mapping degrees:

Clearly, the effect is most pronounced when we go from the linear to quadratic mapping. This is also reflected in the error values given in the table above. The effect of going from quadratic to cubic degree is less dramatic, but still tangible owing to a more accurate description of the circular boundary.
Next, let's look at the meshes after three global refinements
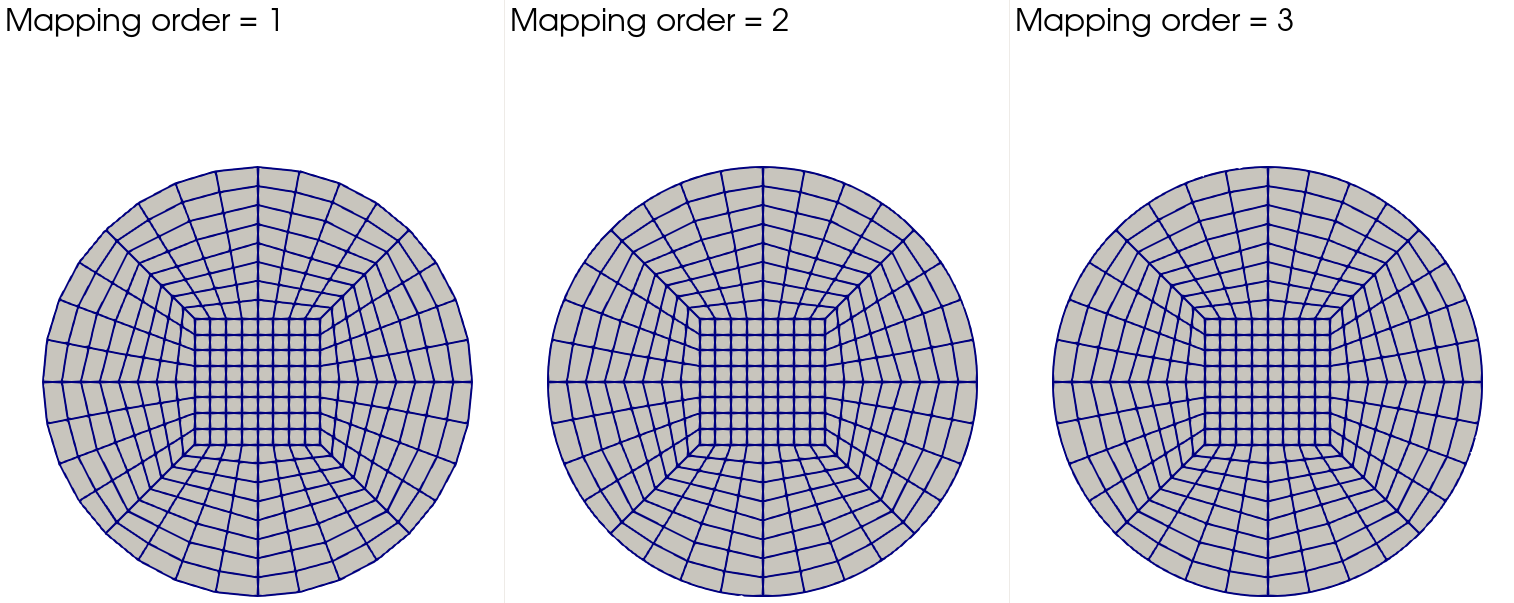
Here, the differences are much less visible, especially for higher order mappings. Indeed, at this refinement level the error values reported in the table are essentially identical between mappings of degrees two and three.
 1.8.17
1.8.17