 |
Reference documentation for deal.II version 9.1.1
|
 |
Reference documentation for deal.II version 9.1.1
|
This tutorial depends on step-20.
This program grew out of a student project by Yan Li at Texas A&M University. Most of the work for this program is by her.
In this project, we propose a numerical simulation for two phase flow problems in porous media. This problem includes one elliptic equation and one nonlinear, time dependent transport equation. This is therefore also the first time-dependent tutorial program (besides the somewhat strange time-dependence of step-18).
The equations covered here are an extension of the material already covered in step-20. In particular, they fall into the class of vector-valued problems. A toplevel overview of this topic can be found in the Handling vector valued problems module.
Modeling of two phase flow in porous media is important for both environmental remediation and the management of petroleum and groundwater reservoirs. Practical situations involving two phase flow include the dispersal of a nonaqueous phase liquid in an aquifer, or the joint movement of a mixture of fluids such as oil and water in a reservoir. Simulation models, if they are to provide realistic predictions, must accurately account for these effects.
To derive the governing equations, consider two phase flow in a reservoir \Omega under the assumption that the movement of fluids is dominated by viscous effects; i.e. we neglect the effects of gravity, compressibility, and capillary pressure. Porosity will be considered to be constant. We will denote variables referring to either of the two phases using subscripts w and o, short for water and oil. The derivation of the equations holds for other pairs of fluids as well, however.
The velocity with which molecules of each of the two phases move is determined by Darcy's law that states that the velocity is proportional to the pressure gradient:
\begin{eqnarray*} \mathbf{u}_{j} = -\frac{k_{rj}(S)}{\mu_{j}} \mathbf{K} \cdot \nabla p \end{eqnarray*}
where \mathbf{u}_{j} is the velocity of phase j=o,w, K is the permeability tensor, k_{rj} is the relative permeability of phase j, p is the pressure and \mu_{j} is the viscosity of phase j. Finally, S is the saturation (volume fraction), i.e. a function with values between 0 and 1 indicating the composition of the mixture of fluids. In general, the coefficients K, k_{rj}, \mu may be spatially dependent variables, and we will always treat them as non-constant functions in the following.
We combine Darcy's law with the statement of conservation of mass for each phase,
\textrm{div}\ \mathbf{u}_{j} = q_j,
with a source term for each phase. By summing over the two phases, we can express the governing equations in terms of the so-called pressure equation:
\begin{eqnarray*} - \nabla \cdot (\mathbf{K}\lambda(S) \nabla p)= q. \end{eqnarray*}
Here, q is the sum source term, and
\lambda(S) = \frac{k_{rw}(S)}{\mu_{w}}+\frac{k_{ro}(S)}{\mu_{o}}
is the total mobility.
So far, this looks like an ordinary stationary, Poisson-like equation that we can solve right away with the techniques of the first few tutorial programs (take a look at step-6, for example, for something very similar). However, we have not said anything yet about the saturation, which of course is going to change as the fluids move around.
The second part of the equations is a therefore description of the dynamics of the saturation. We model this as an advected quantity:
\begin{eqnarray*} S_{t} + \mathbf{u} \cdot \nabla F(S) = 0, \end{eqnarray*}
where \mathbf u is the total velocity
\mathbf{u} = \mathbf{u}_{o} + \mathbf{u}_{w} = -\lambda(S) \mathbf{K}\cdot\nabla p.
In addition,
F(S) = \frac{k_{rw}(S)/\mu_{w}}{k_{rw}(S)/\mu_{w} + k_{ro}(S)/\mu_{o}}
Note that the advection equation contains the term \mathbf{u} \cdot \nabla F(S) rather than \mathbf{u} \cdot \nabla S to indicate that the saturation is not simply transported along; rather, since the two phases move with different velocities, the saturation can actually change even in the advected coordinate system. To see this, rewrite \mathbf{u} \cdot \nabla F(S) = \mathbf{u} F'(S) \cdot \nabla S to observe that the actual velocity with which the phase with saturation S is transported is \mathbf u F'(S) whereas the other phase is transported at velocity \mathbf u (1-F'(S)). F(S) is consequently often referred to as the fractional flow.
In summary, what we get are the following two equations:
\begin{eqnarray*} - \nabla \cdot (\mathbf{K}\lambda(S) \nabla p) &=& q \qquad \textrm{in}\ \Omega\times[0,T], \\ S_{t} + \mathbf{u} \cdot \nabla F(S) &=& 0 \qquad \textrm{in}\ \Omega\times[0,T]. \end{eqnarray*}
Here, p=p(\mathbf x, t), S=S(\mathbf x, t) are now time dependent functions: while at every time instant the flow field is in equilibrium with the pressure (i.e. we neglect dynamic accelerations), the saturation is transported along with the flow and therefore changes over time, in turn affected the flow field again through the dependence of the first equation on S.
This set of equations has a peculiar character: one of the two equations has a time derivative, the other one doesn't. This corresponds to the character that the pressure and velocities are coupled through an instantaneous constraint, whereas the saturation evolves over finite time scales.
Such systems of equations are called Differential Algebraic Equations (DAEs), since one of the equations is a differential equation, the other is not (at least not with respect to the time variable) and is therefore an "algebraic" equation. (The notation comes from the field of ordinary differential equations, where everything that does not have derivatives with respect to the time variable is necessarily an algebraic equation.) This class of equations contains pretty well-known cases: for example, the time dependent Stokes and Navier-Stokes equations (where the algebraic constraint is that the divergence of the flow field, \textrm{div}\ \mathbf u, must be zero) as well as the time dependent Maxwell equations (here, the algebraic constraint is that the divergence of the electric displacement field equals the charge density, \textrm{div}\ \mathbf D = \rho and that the divergence of the magnetic flux density is zero: \textrm{div}\ \mathbf B = 0); even the quasistatic model of step-18 falls into this category. We will see that the different character of the two equations will inform our discretization strategy for the two equations.
In the reservoir simulation community, it is common to solve the equations derived above by going back to the first order, mixed formulation. To this end, we re-introduce the total velocity \mathbf u and write the equations in the following form:
\begin{eqnarray*} \mathbf{u}+\mathbf{K}\lambda(S) \nabla p&=&0 \\ \nabla \cdot\mathbf{u} &=& q \\ S_{t} + \mathbf{u} \cdot \nabla F(S) &=& 0. \end{eqnarray*}
This formulation has the additional benefit that we do not have to express the total velocity \mathbf u appearing in the transport equation as a function of the pressure, but can rather take the primary variable for it. Given the saddle point structure of the first two equations and their similarity to the mixed Laplace formulation we have introduced in step-20, it will come as no surprise that we will use a mixed discretization again.
But let's postpone this for a moment. The first business we have with these equations is to think about the time discretization. In reservoir simulation, there is a rather standard algorithm that we will use here. It first solves the pressure using an implicit equation, then the saturation using an explicit time stepping scheme. The algorithm is called IMPES for IMplicit Pressure Explicit Saturation and was first proposed a long time ago: by Sheldon et al. in 1959 and Stone and Gardner in 1961 (J. W. Sheldon, B. Zondek and W. T. Cardwell: One-dimensional, incompressible, non-capillary, two-phase fluid flow in a porous medium, Trans. SPE AIME, 216 (1959), pp. 290-296; H. L. Stone and A. O. Gardner Jr: Analysis of gas-cap or dissolved-gas reservoirs, Trans. SPE AIME, 222 (1961), pp. 92-104). In a slightly modified form, this algorithm can be written as follows: for each time step, solve
\begin{eqnarray*} \mathbf{u}^{n+1}+\mathbf{K}\lambda(S^n) \nabla p^{n+1}&=&0 \\ \nabla \cdot\mathbf{u}^{n+1} &=& q^{n+1} \\ \frac {S^{n+1}-S^n}{\triangle t} + \mathbf{u}^{n+1} \cdot \nabla F(S^n) &=& 0, \end{eqnarray*}
where \triangle t is the length of a time step. Note how we solve the implicit pressure-velocity system that only depends on the previously computed saturation S^n, and then do an explicit time step for S^{n+1} that only depends on the previously known S^n and the just computed \mathbf{u}^{n+1}. This way, we never have to iterate for the nonlinearities of the system as we would have if we used a fully implicit method.
We can then state the problem in weak form as follows, by multiplying each equation with test functions \mathbf v, \phi, and \sigma and integrating terms by parts:
\begin{eqnarray*} \left((\mathbf{K}\lambda(S^n))^{-1} \mathbf{u}^{n+1},\mathbf v\right)_\Omega - (p^{n+1}, \nabla\cdot\mathbf v)_\Omega &=& - (p^{n+1}, \mathbf v)_{\partial\Omega} \\ (\nabla \cdot\mathbf{u}^{n+1}, \phi)_\Omega &=& (q^{n+1},\phi)_\Omega \end{eqnarray*}
Note that in the first term, we have to prescribe the pressure p^{n+1} on the boundary \partial\Omega as boundary values for our problem. \mathbf n denotes the unit outward normal vector to \partial K, as usual.
For the saturation equation, we obtain after integrating by parts
\begin{eqnarray*} (S^{n+1}, \sigma)_\Omega - \triangle t \sum_K \left\{ \left(F(S^n), \nabla \cdot (\mathbf{u}^{n+1} \sigma)\right)_K - \left(F(S^n) (\mathbf n \cdot \mathbf{u}^{n+1}, \sigma\right)_{\partial K} \right\} &=& (S^n,\sigma)_\Omega. \end{eqnarray*}
Using the fact that \nabla \cdot \mathbf{u}^{n+1}=q^{n+1}, we can rewrite the cell term to get an equation as follows:
\begin{eqnarray*} (S^{n+1}, \sigma)_\Omega - \triangle t \sum_K \left\{ \left(F(S^n) \mathbf{u}^{n+1}, \nabla \sigma\right)_K - \left(F(S^n) (\mathbf n \cdot \mathbf{u}^{n+1}), \sigma\right)_{\partial K} \right\} &=& (S^n,\sigma)_\Omega + \triangle t \sum_K \left(F(S^n) q^{n+1}, \sigma\right)_K. \end{eqnarray*}
In each time step, we then apply the mixed finite method of step-20 to the velocity and pressure. To be well-posed, we choose Raviart-Thomas spaces RT_{k} for \mathbf{u} and discontinuous elements of class DGQ_{k} for p. For the saturation, we will also choose DGQ_{k} spaces.
Since we have discontinuous spaces, we have to think about how to evaluate terms on the interfaces between cells, since discontinuous functions are not really defined there. In particular, we have to give a meaning to the last term on the left hand side of the saturation equation. To this end, let us define that we want to evaluate it in the following sense:
\begin{eqnarray*} &&\left(F(S^n) (\mathbf n \cdot \mathbf{u}^{n+1}), \sigma\right)_{\partial K} \\ &&\qquad = \left(F(S^n_+) (\mathbf n \cdot \mathbf{u}^{n+1}_+), \sigma\right)_{\partial K_+} + \left(F(S^n_-) (\mathbf n \cdot \mathbf{u}^{n+1}_-), \sigma\right)_{\partial K_-}, \end{eqnarray*}
where \partial K_{-} \dealcoloneq \{x\in \partial K, \mathbf{u}(x) \cdot \mathbf{n}<0\} denotes the inflow boundary and \partial K_{+} \dealcoloneq \{\partial K \setminus \partial K_{-}\} is the outflow part of the boundary. The quantities S_+,\mathbf{u}_+ then correspond to the values of these variables on the present cell, whereas S_-,\mathbf{u}_- (needed on the inflow part of the boundary of K) are quantities taken from the neighboring cell. Some more context on discontinuous element techniques and evaluation of fluxes can also be found in step-12.
The linear solvers used in this program are a straightforward extension of the ones used in step-20 (but without LinearOperator). Essentially, we simply have to extend everything from two to three solution components. If we use the discrete spaces mentioned above and put shape functions into the bilinear forms, we arrive at the following linear system to be solved for time step n+1:
\left( \begin{array}{ccc} M^u(S^{n}) & B^{T}& 0\\ B & 0 & 0\\ \triangle t\; H & 0& M^S \end{array} \right) \left( \begin{array}{c} \mathbf{U}^{n+1} \\ P^{n+1} \\ S^{n+1} \end{array} \right) = \left( \begin{array}{c} 0 \\ F_2 \\ F_3 \end{array} \right)
where the individual matrices and vectors are defined as follows using shape functions \mathbf v_i (of type Raviart Thomas RT_k) for velocities and \phi_i (of type DGQ_k) for both pressures and saturations:
\begin{eqnarray*} M^u(S^n)_{ij} &=& \left((\mathbf{K}\lambda(S^n))^{-1} \mathbf{v}_i,\mathbf v_j\right)_\Omega, \\ B_{ij} &=& -(\nabla \cdot \mathbf v_j, \phi_i)_\Omega, \\ H_{ij} &=& - \sum_K \left\{ \left(F(S^n) \mathbf v_i, \nabla \phi_j)\right)_K - \left(F(S^n_+) (\mathbf n \cdot (\mathbf v_i)_+), \phi_j\right)_{\partial K_+} - \left(F(S^n_-) (\mathbf n \cdot (\mathbf v_i)_-), \phi_j\right)_{\partial K_-}, \right\} \\ M^S_{ij} &=& (\phi_i, \phi_j)_\Omega, \\ (F_2)_i &=& -(q^{n+1},\phi_i)_\Omega, \\ (F_3)_i &=& (S^n,\phi_i)_\Omega +\triangle t \sum_K \left(F(S^n) q^{n+1}, \phi_i\right)_K. \end{eqnarray*}
The system above presents a complication: Since the matrix H_{ij} depends on \mathbf u^{n+1} implicitly (the velocities are needed to determine which parts of the boundaries \partial K of cells are influx or outflux parts), we can only assemble this matrix after we have solved for the velocities.
The solution scheme then involves the following steps:
Solve for the pressure p^{n+1} using the Schur complement technique introduced in step-20.
Solve for the velocity \mathbf u^{n+1} as also discussed in step-20.
Compute the term F_3-\triangle t\; H \mathbf u^{n+1}, using the just computed velocities.
In this scheme, we never actually build the matrix H, but rather generate the right hand side of the third equation once we are ready to do so.
In the program, we use a variable solution to store the solution of the present time step. At the end of each step, we copy its content, i.e. all three of its block components, into the variable old_solution for use in the next time step.
A general rule of thumb in hyperbolic transport equations like the equation we have to solve for the saturation equation is that if we use an explicit time stepping scheme, then we should use a time step such that the distance that a particle can travel within one time step is no larger than the diameter of a single cell. In other words, here, we should choose
\triangle t_{n+1} \le \frac h{|\mathbf{u}^{n+1}(\mathbf{x})|}.
Fortunately, we are in a position where we can do that: we only need the time step when we want to assemble the right hand side of the saturation equation, which is after we have already solved for \mathbf{u}^{n+1}. All we therefore have to do after solving for the velocity is to loop over all quadrature points in the domain and determine the maximal magnitude of the velocity. We can then set the time step for the saturation equation to
\triangle t_{n+1} = \frac {\min_K h_K}{\max_{\mathbf{x}}|\mathbf{u}^{n+1}(\mathbf{x})|}.
Why is it important to do this? If we don't, then we will end up with lots of places where our saturation is larger than one or less than zero, as can easily be verified. (Remember that the saturation corresponds to something like the water fraction in the fluid mixture, and therefore must physically be between 0 and 1.) On the other hand, if we choose our time step according to the criterion listed above, this only happens very very infrequently — in fact only once for the entire run of the program. However, to be on the safe side, however, we run a function project_back_saturation at the end of each time step, that simply projects the saturation back onto the interval [0,1], should it have gotten out of the physical range. This is useful since the functions \lambda(S) and F(S) do not represent anything physical outside this range, and we should not expect the program to do anything useful once we have negative saturations or ones larger than one.
Note that we will have similar restrictions on the time step also in step-23 and step-24 where we solve the time dependent wave equation, another hyperbolic problem. We will also come back to the issue of time step choice below in the section on possible extensions to this program.
For simplicity, this program assumes that there is no source, q=0, and that the heterogeneous porous medium is isotropic \mathbf{K}(\mathbf{x}) = k(\mathbf{x}) \mathbf{I}. The first one of these is a realistic assumption in oil reservoirs: apart from injection and production wells, there are usually no mechanisms for fluids to appear or disappear out of the blue. The second one is harder to justify: on a microscopic level, most rocks are isotropic, because they consist of a network of interconnected pores. However, this microscopic scale is out of the range of today's computer simulations, and we have to be content with simulating things on the scale of meters. On that scale, however, fluid transport typically happens through a network of cracks in the rock, rather than through pores. However, cracks often result from external stress fields in the rock layer (for example from tectonic faulting) and the cracks are therefore roughly aligned. This leads to a situation where the permeability is often orders of magnitude larger in the direction parallel to the cracks than perpendicular to the cracks. A problem typically faces in reservoir simulation, however, is that the modeler doesn't know the direction of cracks because oil reservoirs are not accessible to easy inspection. The only solution in that case is to assume an effective, isotropic permeability.
Whatever the matter, both of these restrictions, no sources and isotropy, would be easy to lift with a few lines of code in the program.
Next, for simplicity, our numerical simulation will be done on the unit cell \Omega = [0,1]\times [0,1] for t\in [0,T]. Our initial conditions are S(\mathbf{x},0)=0; in the oil reservoir picture, where S would indicate the water saturation, this means that the reservoir contains pure oil at the beginning. Note that we do not need any initial conditions for pressure or velocity, since the equations do not contain time derivatives of these variables. Finally, we impose the following pressure boundary conditions:
p(\mathbf{x},t)=1-x_1 \qquad \textrm{on}\ \partial\Omega.
Since the pressure and velocity solve a mixed form Poisson equation, the imposed pressure leads to a resulting flow field for the velocity. On the other hand, this flow field determines whether a piece of the boundary is of inflow or outflow type, which is of relevance because we have to impose boundary conditions for the saturation on the inflow part of the boundary,
\Gamma_{in}(t) = \{\mathbf{x}\in\partial\Omega: \mathbf{n} \cdot \mathbf{u}(\mathbf{x},t) < 0\}.
On this inflow boundary, we impose the following saturation values:
\begin{eqnarray} S(\mathbf{x},t) = 1 & \textrm{on}\ \Gamma_{in}\cap\{x_1=0\}, \\ S(\mathbf{x},t) = 0 & \textrm{on}\ \Gamma_{in}\backslash \{x_1=0\}. \end{eqnarray}
In other words, we have pure water entering the reservoir at the left, whereas the other parts of the boundary are in contact with undisturbed parts of the reservoir and whenever influx occurs on these boundaries, pure oil will enter.
In our simulations, we choose the total mobility as
\lambda (S) = \frac{1.0}{\mu} S^2 +(1-S)^2
where we use \mu=0.2 for the viscosity. In addition, the fractional flow of water is given by
F(S)=\frac{S^2}{S^2+\mu (1-S)^2}
Finally, to come back to the description of the testcase, we will show results for computations with the two permeability functions introduced at the end of the results section of step-20:
A function that models a single, winding crack that snakes through the domain. In analogy to step-20, but taking care of the slightly different geometry we have here, we describe this by the following function:
k(\mathbf x) = \max \left\{ e^{-\left(\frac{x_2-\frac 12 - 0.1\sin(10x_1)}{0.1}\right)^2}, 0.01 \right\}.
Taking the maximum is necessary to ensure that the ratio between maximal and minimal permeability remains bounded. If we don't do that, permeabilities will span many orders of magnitude. On the other hand, the ratio between maximal and minimal permeability is a factor in the condition number of the Schur complement matrix, and if too large leads to problems for which our linear solvers will no longer converge properly.
\begin{eqnarray*} k(\mathbf x) &=& \min \left\{ \max \left\{ \sum_{i=1}^N \sigma_i(\mathbf{x}), 0.01 \right\}, 4\right\}, \\ \sigma_i(\mathbf x) &=& e^{-\left(\frac{|\mathbf{x}-\mathbf{x}_i|}{0.05}\right)^2}, \end{eqnarray*}
where the centers \mathbf{x}_i are N randomly chosen locations inside the domain. This function models a domain in which there are N centers of higher permeability (for example where rock has cracked) embedded in a matrix of more pristine, unperturbed background rock. Note that here we have cut off the permeability function both above and below to ensure a bounded condition number.This program is an adaptation of step-20 and includes some technique of DG methods from step-12. A good part of the program is therefore very similar to step-20 and we will not comment again on these parts. Only the new stuff will be discussed in more detail.
All of these include files have been used before:
In this program, we use a tensor-valued coefficient. Since it may have a spatial dependence, we consider it a tensor-valued function. The following include file provides the TensorFunction class that offers such functionality:
The last step is as in all previous programs:
TwoPhaseFlowProblem classThis is the main class of the program. It is close to the one of step-20, but with a few additional functions:
assemble_rhs_S assembles the right hand side of the saturation equation. As explained in the introduction, this can't be integrated into assemble_rhs since it depends on the velocity that is computed in the first part of the time step.
get_maximal_velocity does as its name suggests. This function is used in the computation of the time step size.
project_back_saturation resets all saturation degrees of freedom with values less than zero to zero, and all those with saturations greater than one to one. The rest of the class should be pretty much obvious. The viscosity variable stores the viscosity \mu that enters several of the formulas in the nonlinear equations.
At present, the right hand side of the pressure equation is simply the zero function. However, the rest of the program is fully equipped to deal with anything else, if this is desired:
The next are pressure boundary values. As mentioned in the introduction, we choose a linear pressure field:
Then we also need boundary values on the inflow portions of the boundary. The question whether something is an inflow part is decided when assembling the right hand side, we only have to provide a functional description of the boundary values. This is as explained in the introduction:
Finally, we need initial data. In reality, we only need initial data for the saturation, but we are lazy, so we will later, before the first time step, simply interpolate the entire solution for the previous time step from a function that contains all vector components.
We therefore simply create a function that returns zero in all components. We do that by simply forward every function to the Functions::ZeroFunction class. Why not use that right away in the places of this program where we presently use the InitialValues class? Because this way it is simpler to later go back and choose a different function for initial values.
As announced in the introduction, we implement two different permeability tensor fields. Each of them we put into a namespace of its own, so that it will be easy later to replace use of one by the other in the code.
The first function for the permeability was the one that models a single curving crack. It was already used at the end of step-20, and its functional form is given in the introduction of the present tutorial program. As in some previous programs, we have to declare a (seemingly unnecessary) default constructor of the KInverse class to avoid warnings from some compilers:
This function does as announced in the introduction, i.e. it creates an overlay of exponentials at random places. There is one thing worth considering for this class. The issue centers around the problem that the class creates the centers of the exponentials using a random function. If we therefore created the centers each time we create an object of the present type, we would get a different list of centers each time. That's not what we expect from classes of this type: they should reliably represent the same function.
The solution to this problem is to make the list of centers a static member variable of this class, i.e. there exists exactly one such variable for the entire program, rather than for each object of this type. That's exactly what we are going to do.
The next problem, however, is that we need a way to initialize this variable. Since this variable is initialized at the beginning of the program, we can't use a regular member function for that since there may not be an object of this type around at the time. The C++ standard therefore says that only non-member and static member functions can be used to initialize a static variable. We use the latter possibility by defining a function get_centers that computes the list of center points when called.
Note that this class works just fine in both 2d and 3d, with the only difference being that we use more points in 3d: by experimenting we find that we need more exponentials in 3d than in 2d (we have more ground to cover, after all, if we want to keep the distance between centers roughly equal), so we choose 40 in 2d and 100 in 3d. For any other dimension, the function does presently not know what to do so simply throws an exception indicating exactly this.
There are two more pieces of data that we need to describe, namely the inverse mobility function and the saturation curve. Their form is also given in the introduction:
The linear solvers we use are also completely analogous to the ones used in step-20. The following classes are therefore copied verbatim from there. Note that the classes here are not only copied from step-20, but also duplicate classes in deal.II. In a future version of this example, they should be replaced by an efficient method, though. There is a single change: if the size of a linear system is small, i.e. when the mesh is very coarse, then it is sometimes not sufficient to set a maximum of src.size() CG iterations before the solver in the vmult() function converges. (This is, of course, a result of numerical round-off, since we know that on paper, the CG method converges in at most src.size() steps.) As a consequence, we set the maximum number of iterations equal to the maximum of the size of the linear system and 200.
TwoPhaseFlowProblem class implementationHere now the implementation of the main class. Much of it is actually copied from step-20, so we won't comment on it in much detail. You should try to get familiar with that program first, then most of what is happening here should be mostly clear.
First for the constructor. We use RT_k \times DQ_k \times DQ_k spaces. The time step is set to zero initially, but will be computed before it is needed first, as described in a subsection of the introduction.
This next function starts out with well-known functions calls that create and refine a mesh, and then associate degrees of freedom with it. It does all the same things as in step-20, just now for three components instead of two.
This is the function that assembles the linear system, or at least everything except the (1,3) block that depends on the still-unknown velocity computed during this time step (we deal with this in assemble_rhs_S). Much of it is again as in step-20, but we have to deal with some nonlinearity this time. However, the top of the function is pretty much as usual (note that we set matrix and right hand side to zero at the beginning — something we didn't have to do for stationary problems since there we use each matrix object only once and it is empty at the beginning anyway).
Note that in its present form, the function uses the permeability implemented in the RandomMedium::KInverse class. Switching to the single curved crack permeability function is as simple as just changing the namespace name.
Here's the first significant difference: We have to get the values of the saturation function of the previous time step at the quadrature points. To this end, we can use the FEValues::get_function_values (previously already used in step-9, step-14 and step-15), a function that takes a solution vector and returns a list of function values at the quadrature points of the present cell. In fact, it returns the complete vector-valued solution at each quadrature point, i.e. not only the saturation but also the velocities and pressure:
Then we also have to get the values of the pressure right hand side and of the inverse permeability tensor at the quadrature points:
With all this, we can now loop over all the quadrature points and shape functions on this cell and assemble those parts of the matrix and right hand side that we deal with in this function. The individual terms in the contributions should be self-explanatory given the explicit form of the bilinear form stated in the introduction:
Next, we also have to deal with the pressure boundary values. This, again is as in step-20:
The final step in the loop over all cells is to transfer local contributions into the global matrix and right hand side vector:
So much for assembly of matrix and right hand side. Note that we do not have to interpolate and apply boundary values since they have all been taken care of in the weak form already.
As explained in the introduction, we can only evaluate the right hand side of the saturation equation once the velocity has been computed. We therefore have this separate function to this end.
First for the cell terms. These are, following the formulas in the introduction, (S^n,\sigma)-(F(S^n) \mathbf{v}^{n+1},\nabla \sigma), where \sigma is the saturation component of the test function:
Secondly, we have to deal with the flux parts on the face boundaries. This was a bit more involved because we first have to determine which are the influx and outflux parts of the cell boundary. If we have an influx boundary, we need to evaluate the saturation on the other side of the face (or the boundary values, if we are at the boundary of the domain).
All this is a bit tricky, but has been explained in some detail already in step-9. Take a look there how this is supposed to work!
After all these preparations, we finally solve the linear system for velocity and pressure in the same way as in step-20. After that, we have to deal with the saturation equation (see below):
First the pressure, using the pressure Schur complement of the first two equations:
Now the velocity:
Finally, we have to take care of the saturation equation. The first business we have here is to determine the time step using the formula in the introduction. Knowing the shape of our domain and that we created the mesh by regular subdivision of cells, we can compute the diameter of each of our cells quite easily (in fact we use the linear extensions in coordinate directions of the cells, not the diameter). Note that we will learn a more general way to do this in step-24, where we use the GridTools::minimal_cell_diameter function.
The maximal velocity we compute using a helper function to compute the maximal velocity defined below, and with all this we can evaluate our new time step length:
The next step is to assemble the right hand side, and then to pass everything on for solution. At the end, we project back saturations onto the physically reasonable range:
There is nothing surprising here. Since the program will do a lot of time steps, we create an output file only every fifth time step and skip all other time steps at the top of the file already.
When creating file names for output close to the bottom of the function, we convert the number of the time step to a string representation that is padded by leading zeros to four digits. We do this because this way all output file names have the same length, and consequently sort well when creating a directory listing.
In this function, we simply run over all saturation degrees of freedom and make sure that if they should have left the physically reasonable range, that they be reset to the interval [0,1]. To do this, we only have to loop over all saturation components of the solution vector; these are stored in the block 2 (block 0 are the velocities, block 1 are the pressures).
It may be instructive to note that this function almost never triggers when the time step is chosen as mentioned in the introduction. However, if we choose the timestep only slightly larger, we get plenty of values outside the proper range. Strictly speaking, the function is therefore unnecessary if we choose the time step small enough. In a sense, the function is therefore only a safety device to avoid situations where our entire solution becomes unphysical because individual degrees of freedom have become unphysical a few time steps earlier.
The following function is used in determining the maximal allowable time step. What it does is to loop over all quadrature points in the domain and find what the maximal magnitude of the velocity is.
This is the final function of our main class. Its brevity speaks for itself. There are only two points worth noting: First, the function projects the initial values onto the finite element space at the beginning; the VectorTools::project function doing this requires an argument indicating the hanging node constraints. We have none in this program (we compute on a uniformly refined mesh), but the function requires the argument anyway, of course. So we have to create a constraint object. In its original state, constraint objects are unsorted, and have to be sorted (using the AffineConstraints::close function) before they can be used. This is what we do here, and which is why we can't simply call the VectorTools::project function with an anonymous temporary object AffineConstraints<double>() as the second argument.
The second point worth mentioning is that we only compute the length of the present time step in the middle of solving the linear system corresponding to each time step. We can therefore output the present end time of a time step only at the end of the time step.
The function as it is here does actually not compute the results found on the web page. The reason is, that even on a decent computer it runs more than a day. If you want to reproduce these results, set the final time at the end of the do loop to 250.
main functionThat's it. In the main function, we pass the degree of the finite element space to the constructor of the TwoPhaseFlowProblem object. Here, we use zero-th degree elements, i.e. RT_0\times DQ_0 \times DQ_0. The rest is as in all the other programs.
If we run the program, we get the following kind of output:
As we can see, the time step is pretty much constant right from the start, which indicates that the velocities in the domain are not strongly dependent on changes in saturation, although they certainly are through the factor \lambda(S) in the pressure equation.
Our second observation is that the number of CG iterations needed to solve the pressure Schur complement equation drops from 22 to 17 between the first and the second time step (in fact, it remains around 17 for the rest of the computations). The reason is actually simple: Before we solve for the pressure during a time step, we don't reset the solution variable to zero. The pressure (and the other variables) therefore have the previous time step's values at the time we get into the CG solver. Since the velocities and pressures don't change very much as computations progress, the previous time step's pressure is actually a good initial guess for this time step's pressure. Consequently, the number of iterations we need once we have computed the pressure once is significantly reduced.
The final observation concerns the number of iterations needed to solve for the saturation, i.e. one. This shouldn't surprise us too much: the matrix we have to solve with is the mass matrix. However, this is the mass matrix for the DGQ_0 element of piecewise constants where no element couples with the degrees of freedom on neighboring cells. The matrix is therefore a diagonal one, and it is clear that we should be able to invert this matrix in a single CG iteration.
With all this, here are a few movies that show how the saturation progresses over time. First, this is for the single crack model, as implemented in the SingleCurvingCrack::KInverse class:

As can be seen, the water rich fluid snakes its way mostly along the high-permeability zone in the middle of the domain, whereas the rest of the domain is mostly impermeable. This and the next movie are generated using n_refinement_steps=7, leading to a 128\times 128 mesh with some 16,000 cells and about 66,000 unknowns in total.
The second movie shows the saturation for the random medium model of class RandomMedium::KInverse, where we have randomly distributed centers of high permeability and fluid hops from one of these zones to the next:
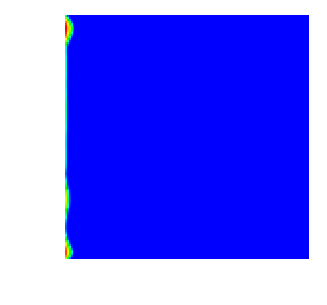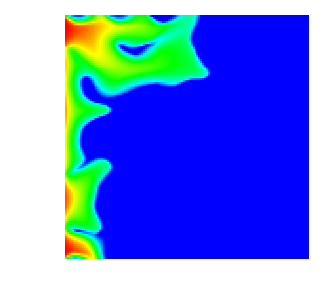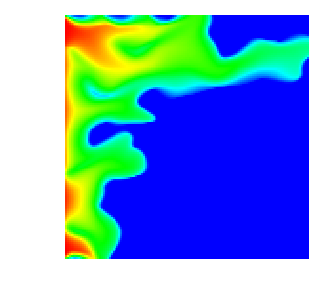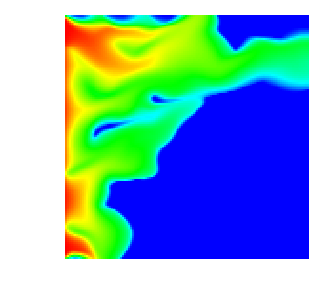
Finally, here is the same situation in three space dimensions, on a mesh with n_refinement_steps=5, which produces a mesh of some 32,000 cells and 167,000 degrees of freedom:

To repeat these computations, all you have to do is to change the line
in the main function to
The visualization uses a cloud technique, where the saturation is indicated by colored but transparent clouds for each cell. This way, one can also see somewhat what happens deep inside the domain. A different way of visualizing would have been to show isosurfaces of the saturation evolving over time. There are techniques to plot isosurfaces transparently, so that one can see several of them at the same time like the layers of an onion.
So why don't we show such isosurfaces? The problem lies in the way isosurfaces are computed: they require that the field to be visualized is continuous, so that the isosurfaces can be generated by following contours at least across a single cell. However, our saturation field is piecewise constant and discontinuous. If we wanted to plot an isosurface for a saturation S=0.5, chances would be that there is no single point in the domain where that saturation is actually attained. If we had to define isosurfaces in that context at all, we would have to take the interfaces between cells, where one of the two adjacent cells has a saturation greater than and the other cell a saturation less than 0.5. However, it appears that most visualization programs are not equipped to do this kind of transformation.
There are a number of areas where this program can be improved. Three of them are listed below. All of them are, in fact, addressed in a tutorial program that forms the continuation of the current one: step-43.
At present, the program is not particularly fast: the 2d random medium computation took about a day for the 1,000 or so time steps. The corresponding 3d computation took almost two days for 800 time steps. The reason why it isn't faster than this is twofold. First, we rebuild the entire matrix in every time step, although some parts such as the B, B^T, and M^S blocks never change.
Second, we could do a lot better with the solver and preconditioners. Presently, we solve the Schur complement B^TM^u(S)^{-1}B with a CG method, using [B^T (\textrm{diag}(M^u(S)))^{-1} B]^{-1} as a preconditioner. Applying this preconditioner is expensive, since it involves solving a linear system each time. This may have been appropriate for step-20, where we have to solve the entire problem only once. However, here we have to solve it hundreds of times, and in such cases it is worth considering a preconditioner that is more expensive to set up the first time, but cheaper to apply later on.
One possibility would be to realize that the matrix we use as preconditioner, B^T (\textrm{diag}(M^u(S)))^{-1} B is still sparse, and symmetric on top of that. If one looks at the flow field evolve over time, we also see that while S changes significantly over time, the pressure hardly does and consequently B^T (\textrm{diag}(M^u(S)))^{-1} B \approx B^T (\textrm{diag}(M^u(S^0)))^{-1} B. In other words, the matrix for the first time step should be a good preconditioner also for all later time steps. With a bit of back-and-forthing, it isn't hard to actually get a representation of it as a SparseMatrix object. We could then hand it off to the SparseMIC class to form a sparse incomplete Cholesky decomposition. To form this decomposition is expensive, but we have to do it only once in the first time step, and can then use it as a cheap preconditioner in the future. We could do better even by using the SparseDirectUMFPACK class that produces not only an incomplete, but a complete decomposition of the matrix, which should yield an even better preconditioner.
Finally, why use the approximation B^T (\textrm{diag}(M^u(S)))^{-1} B to precondition B^T M^u(S)^{-1} B? The latter matrix, after all, is the mixed form of the Laplace operator on the pressure space, for which we use linear elements. We could therefore build a separate matrix A^p on the side that directly corresponds to the non-mixed formulation of the Laplacian, for example using the bilinear form (\mathbf{K}\lambda(S^n) \nabla \varphi_i,\nabla\varphi_j). We could then form an incomplete or complete decomposition of this non-mixed matrix and use it as a preconditioner of the mixed form.
Using such techniques, it can reasonably be expected that the solution process will be faster by at least an order of magnitude.
In the introduction we have identified the time step restriction
\triangle t_{n+1} \le \frac h{|\mathbf{u}^{n+1}(\mathbf{x})|}
that has to hold globally, i.e. for all \mathbf x. After discretization, we satisfy it by choosing
\triangle t_{n+1} = \frac {\min_K h_K}{\max_{\mathbf{x}}|\mathbf{u}^{n+1}(\mathbf{x})|}.
This restriction on the time step is somewhat annoying: the finer we make the mesh the smaller the time step; in other words, we get punished twice: each time step is more expensive to solve and we have to do more time steps.
This is particularly annoying since the majority of the additional work is spent solving the implicit part of the equations, i.e. the pressure-velocity system, whereas it is the hyperbolic transport equation for the saturation that imposes the time step restriction.
To avoid this bottleneck, people have invented a number of approaches. For example, they may only re-compute the pressure-velocity field every few time steps (or, if you want, use different time step sizes for the pressure/velocity and saturation equations). This keeps the time step restriction on the cheap explicit part while it makes the solution of the implicit part less frequent. Experiments in this direction are certainly worthwhile; one starting point for such an approach is the paper by Zhangxin Chen, Guanren Huan and Baoyan Li: An improved IMPES method for two-phase flow in porous media, Transport in Porous Media, 54 (2004), pp. 361—376. There are certainly many other papers on this topic as well, but this one happened to land on our desk a while back.
Adaptivity would also clearly help. Looking at the movies, one clearly sees that most of the action is confined to a relatively small part of the domain (this particularly obvious for the saturation, but also holds for the velocities and pressures). Adaptivity can therefore be expected to keep the necessary number of degrees of freedom low, or alternatively increase the accuracy.
On the other hand, adaptivity for time dependent problems is not a trivial thing: we would have to change the mesh every few time steps, and we would have to transport our present solution to the next mesh every time we change it (something that the SolutionTransfer class can help with). These are not insurmountable obstacles, but they do require some additional coding and more than we felt comfortable was worth packing into this tutorial program.
 1.8.14
1.8.14