This tutorial depends on step-31, step-55.
This program was contributed by Martin Kronbichler, Wolfgang Bangerth, and Timo Heister.
This material is based upon work partly supported by the National Science Foundation under Award No. EAR-0426271 and The California Institute of Technology; and in a continuation by the National Science Foundation under Award No. EAR-0949446 and The University of California – Davis. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author and do not necessarily reflect the views of the National Science Foundation, The California Institute of Technology, or of The University of California – Davis.
The work discussed here is also presented in the following publication: M. Kronbichler, T. Heister, W. Bangerth: High Accuracy Mantle Convection Simulation through Modern Numerical Methods, Geophysical Journal International, 2012, 191, 12-29. [DOI]
The continuation of development of this program has led to the much larger open source code ASPECT (see http://aspect.geodynamics.org/) which is much more flexible in solving many kinds of related problems.
Introduction
This program does pretty much exactly what step-31 already does: it solves the Boussinesq equations that describe the motion of a fluid whose temperature is not in equilibrium. As such, all the equations we have described in step-31 still hold: we solve the same general partial differential equation (with only minor modifications to adjust for more realism in the problem setting), using the same finite element scheme, the same time stepping algorithm, and more or less the same stabilization method for the temperature advection-diffusion equation. As a consequence, you may first want to understand that program — and its implementation — before you work on the current one.
The difference between step-31 and the current program is that here we want to do things in parallel, using both the availability of many machines in a cluster (with parallelization based on MPI) as well as many processor cores within a single machine (with parallelization based on threads). This program's main job is therefore to introduce the changes that are necessary to utilize the availability of these parallel compute resources. In this regard, it builds on the step-40 program that first introduces the necessary classes for much of the parallel functionality, and on step-55 that shows how this is done for a vector-valued problem.
In addition to these changes, we also use a slightly different preconditioner, and we will have to make a number of changes that have to do with the fact that we want to solve a realistic problem here, not a model problem. The latter, in particular, will require that we think about scaling issues as well as what all those parameters and coefficients in the equations under consideration actually mean. We will discuss first the issues that affect changes in the mathematical formulation and solver structure, then how to parallelize things, and finally the actual testcase we will consider.
Using the "right" pressure
In step-31, we used the following Stokes model for the velocity and pressure field:
\begin{eqnarray*}
-\nabla \cdot (2 \eta \varepsilon ({\mathbf u})) + \nabla p &=&
-\rho \; \beta \; T \mathbf{g},
\\
\nabla \cdot {\mathbf u} &=& 0.
\end{eqnarray*}
The right hand side of the first equation appears a wee bit unmotivated. Here's how things should really be. We need the external forces that act on the fluid, which we assume are given by gravity only. In the current case, we assume that the fluid does expand slightly for the purposes of this gravity force, but not enough that we need to modify the incompressibility condition (the second equation). What this means is that for the purpose of the right hand side, we can assume that \(\rho=\rho(T)\). An assumption that may not be entirely justified is that we can assume that the changes of density as a function of temperature are small, leading to an expression of the form \(\rho(T) = \rho_{\text{ref}}
[1-\beta(T-T_{\text{ref}})]\), i.e., the density equals \(\rho_{\text{ref}}\) at reference temperature and decreases linearly as the temperature increases (as the material expands). The force balance equation then looks properly written like this:
\begin{eqnarray*}
-\nabla \cdot (2 \eta \varepsilon ({\mathbf u})) + \nabla p &=&
\rho_{\text{ref}} [1-\beta(T-T_{\text{ref}})] \mathbf{g}.
\end{eqnarray*}
Now note that the gravity force results from a gravity potential as \(\mathbf g=-\nabla \varphi\), so that we can re-write this as follows:
\begin{eqnarray*}
-\nabla \cdot (2 \eta \varepsilon ({\mathbf u})) + \nabla p &=&
-\rho_{\text{ref}} \; \beta\; T\; \mathbf{g}
-\rho_{\text{ref}} [1+\beta T_{\text{ref}}] \nabla\varphi.
\end{eqnarray*}
The second term on the right is time independent, and so we could introduce a new "dynamic" pressure \(p_{\text{dyn}}=p+\rho_{\text{ref}}
[1+\beta T_{\text{ref}}] \varphi=p_{\text{total}}-p_{\text{static}}\) with which the Stokes equations would read:
\begin{eqnarray*}
-\nabla \cdot (2 \eta \varepsilon ({\mathbf u})) + \nabla p_{\text{dyn}} &=&
-\rho_{\text{ref}} \; \beta \; T \; \mathbf{g},
\\
\nabla \cdot {\mathbf u} &=& 0.
\end{eqnarray*}
This is exactly the form we used in step-31, and it was appropriate to do so because all changes in the fluid flow are only driven by the dynamic pressure that results from temperature differences. (In other words: Any contribution to the right hand side that results from taking the gradient of a scalar field have no effect on the velocity field.)
On the other hand, we will here use the form of the Stokes equations that considers the total pressure instead:
\begin{eqnarray*}
-\nabla \cdot (2 \eta \varepsilon ({\mathbf u})) + \nabla p &=&
\rho(T)\; \mathbf{g},
\\
\nabla \cdot {\mathbf u} &=& 0.
\end{eqnarray*}
There are several advantages to this:
- This way we can plot the pressure in our program in such a way that it actually shows the total pressure that includes the effects of temperature differences as well as the static pressure of the overlying rocks. Since the pressure does not appear any further in any of the other equations, whether to use one or the other is more a matter of taste than of correctness. The flow field is exactly the same, but we get a pressure that we can now compare with values that are given in geophysical books as those that hold at the bottom of the earth mantle, for example.
- If we wanted to make the model even more realistic, we would have to take into account that many of the material parameters (e.g. the viscosity, the density, etc) not only depend on the temperature but also the total pressure.
- The model above assumed a linear dependence \(\rho(T) = \rho_{\text{ref}}
[1-\beta(T-T_{\text{ref}})]\) and assumed that \(\beta\) is small. In practice, this may not be so. In fact, realistic models are certainly not linear, and \(\beta\) may also not be small for at least part of the temperature range because the density's behavior is substantially dependent not only on thermal expansion but by phase changes.
- A final reason to do this is discussed in the results section and concerns possible extensions to the model we use here. It has to do with the fact that the temperature equation (see below) we use here does not include a term that contains the pressure. It should, however: rock, like gas, heats up as you compress it. Consequently, material that rises up cools adiabatically, and cold material that sinks down heats adiabatically. We discuss this further below.
- Note
- There is, however, a downside to this procedure. In the earth, the dynamic pressure is several orders of magnitude smaller than the total pressure. If we use the equations above and solve all variables to, say, 4 digits of accuracy, then we may be able to get the velocity and the total pressure right, but we will have no accuracy at all if we compute the dynamic pressure by subtracting from the total pressure the static part \(p_\text{static}=\rho_{\text{ref}}
[1+\beta T_{\text{ref}}] \varphi\). If, for example, the dynamic pressure is six orders of magnitude smaller than the static pressure, then we need to solve the overall pressure to at least seven digits of accuracy to get anything remotely accurate. That said, in practice this turns out not to be a limiting factor.
The scaling of discretized equations
Remember that we want to solve the following set of equations:
\begin{eqnarray*}
-\nabla \cdot (2 \eta \varepsilon ({\mathbf u})) + \nabla p &=&
\rho(T) \mathbf{g},
\\
\nabla \cdot {\mathbf u} &=& 0,
\\
\frac{\partial T}{\partial t}
+
{\mathbf u} \cdot \nabla T
-
\nabla \cdot \kappa \nabla T &=& \gamma,
\end{eqnarray*}
augmented by appropriate boundary and initial conditions. As discussed in step-31, we will solve this set of equations by solving for a Stokes problem first in each time step, and then moving the temperature equation forward by one time interval.
The problem under consideration in this current section is with the Stokes problem: if we discretize it as usual, we get a linear system
\begin{eqnarray*}
M \; X
=
\left(\begin{array}{cc}
A & B^T \\ B & 0
\end{array}\right)
\left(\begin{array}{c}
U \\ P
\end{array}\right)
=
\left(\begin{array}{c}
F_U \\ 0
\end{array}\right)
=
F
\end{eqnarray*}
which in this program we will solve with a FGMRES solver. This solver iterates until the residual of these linear equations is below a certain tolerance, i.e., until
\[
\left\|
\left(\begin{array}{c}
F_U - A U^{(k)} - B P^{(k)}
\\
B^T U^{(k)}
\end{array}\right)
\right\|
< \text{Tol}.
\]
This does not make any sense from the viewpoint of physical units: the quantities involved here have physical units so that the first part of the residual has units \(\frac{\text{Pa}}{\text{m}}
\text{m}^{\text{dim}}\) (most easily established by considering the term \((\nabla \cdot \mathbf v, p)_{\Omega}\) and considering that the pressure has units \(\text{Pa}=\frac{\text{kg}}{\text{m}\;\text{s}^2}\) and the integration yields a factor of \(\text{m}^{\text{dim}}\)), whereas the second part of the residual has units \(\frac{\text{m}^{\text{dim}}}{\text{s}}\). Taking the norm of this residual vector would yield a quantity with units \(\text{m}^{\text{dim}-1} \sqrt{\left(\text{Pa}\right)^2 +
\left(\frac{\text{m}}{\text{s}}\right)^2}\). This, quite obviously, does not make sense, and we should not be surprised that doing so is eventually going to come back hurting us.
So why is this an issue here, but not in step-31? The reason back there is that everything was nicely balanced: velocities were on the order of one, the pressure likewise, the viscosity was one, and the domain had a diameter of \(\sqrt{2}\). As a result, while nonsensical, nothing bad happened. On the other hand, as we will explain below, things here will not be that simply scaled: \(\eta\) will be around \(10^{21}\), velocities on the order of \(10^{-8}\), pressure around \(10^8\), and the diameter of the domain is \(10^7\). In other words, the order of magnitude for the first equation is going to be \(\eta\text{div}\varepsilon(\mathbf u) \approx 10^{21} \frac{10^{-8}}{(10^7)^2}
\approx 10^{-1}\), whereas the second equation will be around \(\text{div}{\mathbf u}\approx \frac{10^{-8}}{10^7} \approx 10^{-15}\). Well, so what this will lead to is this: if the solver wants to make the residual small, it will almost entirely focus on the first set of equations because they are so much bigger, and ignore the divergence equation that describes mass conservation. That's exactly what happens: unless we set the tolerance to extremely small values, the resulting flow field is definitely not divergence free. As an auxiliary problem, it turns out that it is difficult to find a tolerance that always works; in practice, one often ends up with a tolerance that requires 30 or 40 iterations for most time steps, and 10,000 for some others.
So what's a numerical analyst to do in a case like this? The answer is to start at the root and first make sure that everything is mathematically consistent first. In our case, this means that if we want to solve the system of Stokes equations jointly, we have to scale them so that they all have the same physical dimensions. In our case, this means multiplying the second equation by something that has units \(\frac{\text{Pa}\;\text{s}}{\text{m}}\); one choice is to multiply with \(\frac{\eta}{L}\) where \(L\) is a typical lengthscale in our domain (which experiments show is best chosen to be the diameter of plumes — around 10 km — rather than the diameter of the domain). Using these numbers for \(\eta\) and \(L\), this factor is around \(10^{17}\). So, we now get this for the Stokes system:
\begin{eqnarray*}
-\nabla \cdot (2 \eta \varepsilon ({\mathbf u})) + \nabla p &=&
\rho(T) \; \mathbf{g},
\\
\frac{\eta}{L} \nabla \cdot {\mathbf u} &=& 0.
\end{eqnarray*}
The trouble with this is that the result is not symmetric any more (we have \(\frac{\eta}{L} \nabla \cdot\) at the bottom left, but not its transpose operator at the top right). This, however, can be cured by introducing a scaled pressure \(\hat p = \frac{L}{\eta}p\), and we get the scaled equations
\begin{eqnarray*}
-\nabla \cdot (2 \eta \varepsilon ({\mathbf u})) +
\nabla \left(\frac{\eta}{L} \hat p\right) &=&
\rho(T) \; \mathbf{g},
\\
\frac{\eta}{L} \nabla \cdot {\mathbf u} &=& 0.
\end{eqnarray*}
This is now symmetric. Obviously, we can easily recover the original pressure \(p\) from the scaled pressure \(\hat p\) that we compute as a result of this procedure.
In the program below, we will introduce a factor EquationData::pressure_scaling that corresponds to \(\frac{\eta}{L}\), and we will use this factor in the assembly of the system matrix and preconditioner. Because it is annoying and error prone, we will recover the unscaled pressure immediately following the solution of the linear system, i.e., the solution vector's pressure component will immediately be unscaled to retrieve the physical pressure. Since the solver uses the fact that we can use a good initial guess by extrapolating the previous solutions, we also have to scale the pressure immediately before solving.
Changes to the Stokes preconditioner and solver
In this tutorial program, we apply a variant of the preconditioner used in step-31. That preconditioner was built to operate on the system matrix \(M\) in block form such that the product matrix
\begin{eqnarray*}
P^{-1} M
=
\left(\begin{array}{cc}
A^{-1} & 0 \\ S^{-1} B A^{-1} & -S^{-1}
\end{array}\right)
\left(\begin{array}{cc}
A & B^T \\ B & 0
\end{array}\right)
\end{eqnarray*}
is of a form that Krylov-based iterative solvers like GMRES can solve in a few iterations. We then replaced the exact inverse of \(A\) by the action of an AMG preconditioner \(\tilde{A}\) based on a vector Laplace matrix, approximated the Schur complement \(S = B A^{-1} B^T\) by a mass matrix \(M_p\) on the pressure space and wrote an InverseMatrix class for implementing the action of \(M_p^{-1}\approx S^{-1}\) on vectors. In the InverseMatrix class, we used a CG solve with an incomplete Cholesky (IC) preconditioner for performing the inner solves.
An observation one can make is that we use just the action of a preconditioner for approximating the velocity inverse \(A^{-1}\) (and the outer GMRES iteration takes care of the approximate character of the inverse), whereas we use a more or less exact inverse for \(M_p^{-1}\), realized by a fully converged CG solve. This appears unbalanced, but there's system to this madness: almost all the effort goes into the upper left block to which we apply the AMG preconditioner, whereas even an exact inversion of the pressure mass matrix costs basically nothing. Consequently, if it helps us reduce the overall number of iterations somewhat, then this effort is well spent.
That said, even though the solver worked well for step-31, we have a problem here that is a bit more complicated (cells are deformed, the pressure varies by orders of magnitude, and we want to plan ahead for more complicated physics), and so we'll change a few things slightly:
- For more complex problems, it turns out that using just a single AMG V-cycle as preconditioner is not always sufficient. The outer solver converges just fine most of the time in a reasonable number of iterations (say, less than 50) but there are the occasional time step where it suddenly takes 700 or so. What exactly is going on there is hard to determine, but the problem can be avoided by using a more accurate solver for the top left block. Consequently, we'll want to use a CG iteration to invert the top left block of the preconditioner matrix, and use the AMG as a preconditioner for the CG solver.
- The downside of this is that, of course, the Stokes preconditioner becomes much more expensive (approximately 10 times more expensive than when we just use a single V-cycle). Our strategy then is this: let's do up to 30 GMRES iterations with just the V-cycle as a preconditioner and if that doesn't yield convergence, then take the best approximation of the Stokes solution obtained after this first round of iterations and use that as the starting guess for iterations where we use the full inner solver with a rather lenient tolerance as preconditioner. In all our experiments this leads to convergence in only a few additional iterations.
- One thing we need to pay attention to is that when using a CG with a lenient tolerance in the preconditioner, then \(y = \tilde A^{-1} r\) is no longer a linear function of \(r\) (it is, of course, if we have a very stringent tolerance in our solver, or if we only apply a single V-cycle). This is a problem since now our preconditioner is no longer a linear operator; in other words, every time GMRES uses it the preconditioner looks different. The standard GMRES solver can't deal with this, leading to slow convergence or even breakdown, but the F-GMRES variant is designed to deal with exactly this kind of situation and we consequently use it.
- On the other hand, once we have settled on using F-GMRES we can relax the tolerance used in inverting the preconditioner for \(S\). In step-31, we ran a preconditioned CG method on \(\tilde S\) until the residual had been reduced by 7 orders of magnitude. Here, we can again be more lenient because we know that the outer preconditioner doesn't suffer.
- In step-31, we used a left preconditioner in which we first invert the top left block of the preconditioner matrix, then apply the bottom left (divergence) one, and then invert the bottom right. In other words, the application of the preconditioner acts as a lower left block triangular matrix. Another option is to use a right preconditioner that here would be upper right block triangulation, i.e., we first invert the bottom right Schur complement, apply the top right (gradient) operator and then invert the elliptic top left block. To a degree, which one to choose is a matter of taste. That said, there is one significant advantage to a right preconditioner in GMRES-type solvers: the residual with which we determine whether we should stop the iteration is the true residual, not the norm of the preconditioned equations. Consequently, it is much simpler to compare it to the stopping criterion we typically use, namely the norm of the right hand side vector. In writing this code we found that the scaling issues we discussed above also made it difficult to determine suitable stopping criteria for left-preconditioned linear systems, and consequently this program uses a right preconditioner.
- In step-31, we used an IC (incomplete Cholesky) preconditioner for the pressure mass matrix in the Schur complement preconditioner and for the solution of the temperature system. Here, we could in principle do the same, but we do choose an even simpler preconditioner, namely a Jacobi preconditioner for both systems. This is because here we target at massively parallel computations, where the decompositions for IC/ILU would have to be performed block-wise for the locally owned degrees of freedom on each processor. This means, that the preconditioner gets more like a Jacobi preconditioner anyway, so we rather start from that variant straight away. Note that we only use the Jacobi preconditioners for CG solvers with mass matrices, where they give optimal (h-independent) convergence anyway, even though they usually require about twice as many iterations as an IC preconditioner.
As a final note, let us remark that in step-31 we computed the Schur complement \(S=B A^{-1} B^T\) by approximating \(-\text{div}(-\eta\Delta)^{-1}\nabla \approx \frac 1{\eta} \mathbf{1}\). Now, however, we have re-scaled the \(B\) and \(B^T\) operators. So \(S\) should now approximate \(-\frac{\eta}{L}\text{div}(-\eta\Delta)^{-1}\nabla \frac{\eta}{L} \approx
\left(\frac{\eta}{L}\right)^2 \frac 1{\eta} \mathbf{1}\). We use the discrete form of the right hand side of this as our approximation \(\tilde S\) to \(S\).
Changes to the artificial viscosity stabilization
Similarly to step-31, we will use an artificial viscosity for stabilization based on a residual of the equation. As a difference to step-31, we will provide two slightly different definitions of the stabilization parameter. For \(\alpha=1\), we use the same definition as in step-31:
\begin{eqnarray*}
\nu_\alpha(T)|_K
=
\nu_1(T)|_K
=
\beta
\|\mathbf{u}\|_{L^\infty(K)}
h_K
\min\left\{
1,
\frac{\|R_1(T)\|_{L^\infty(K)}}{c(\mathbf{u},T)}
\right\}
\end{eqnarray*}
where we compute the viscosity from a residual \(\|R_1(T)\|_{L^\infty(K)}\) of the equation, limited by a diffusion proportional to the mesh size \(h_K\) in regions where the residual is large (around steep gradients). This definition has been shown to work well for the given case, \(\alpha = 1\) in step-31, but it is usually less effective as the diffusion for \(\alpha=2\). For that case, we choose a slightly more readable definition of the viscosity,
\begin{eqnarray*}
\nu_2(T)|_K = \min (\nu_h^\mathrm{max}|_K,\nu_h^\mathrm{E}|_K)
\end{eqnarray*}
where the first term gives again the maximum dissipation (similarly to a first order upwind scheme),
\begin{eqnarray*}
\nu^\mathrm{max}_h|_K = \beta h_K \|\mathbf {u}\|_{L^\infty(K)}
\end{eqnarray*}
and the entropy viscosity is defined as
\begin{eqnarray*}
\nu^\mathrm{E}_h|_K = c_R \frac{h_K^2 \|R_\mathrm{2,E}(T)\|_{L^\infty(K)}}
{\|E(T) - \bar{E}(T)\|_{L^\infty(\Omega)} }.
\end{eqnarray*}
This formula is described in the article J.-L. Guermond, R. Pasquetti, & B. Popov, 2011. Entropy viscosity method for nonlinear conservation laws, J. Comput. Phys., 230, 4248–4267. Compared to the case \(\alpha = 1\), the residual is computed from the temperature entropy, \(E(T) = \frac12 (T-T_m)^2\) with \(T_m\) an average temperature (we choose the mean between the maximum and minimum temperature in the computation), which gives the following formula
\begin{eqnarray*}
R_\mathrm{E}(T) = \frac{\partial E(T)}{\partial t} +
(T-T_\mathrm{m}) \left(\mathbf{u} \cdot \nabla T - \kappa \nabla^2 T - \gamma\right).
\end{eqnarray*}
The denominator in the formula for \(\nu^\mathrm{E}_h|_K\) is computed as the global deviation of the entropy from the space-averaged entropy \(\bar{E}(T) =
\int_\Omega E(T) d\mathbf{x}/\int_\Omega d\mathbf{x}\). As in step-31, we evaluate the artificial viscosity from the temperature and velocity at two previous time levels, in order to avoid a nonlinearity in its definition.
The above definitions of the viscosity are simple, but depend on two parameters, namely \(\beta\) and \(c_R\). For the current program, we want to go about this issue a bit more systematically for both parameters in the case \(\alpha =1\), using the same line of reasoning with which we chose two other parameters in our discretization, \(c_k\) and \(\beta\), in the results section of step-31. In particular, remember that we would like to make the artificial viscosity as small as possible while keeping it as large as necessary. In the following, let us describe the general strategy one may follow. The computations shown here were done with an earlier version of the program and so the actual numerical values you get when running the program may no longer match those shown here; that said, the general approach remains valid and has been used to find the values of the parameters actually used in the program.
To see what is happening, note that below we will impose boundary conditions for the temperature between 973 and 4273 Kelvin, and initial conditions are also chosen in this range; for these considerations, we run the program without internal heat sources or sinks, and consequently the temperature should always be in this range, barring any internal oscillations. If the minimal temperature drops below 973 Kelvin, then we need to add stabilization by either increasing \(\beta\) or decreasing \(c_R\).
As we did in step-31, we first determine an optimal value of \(\beta\) by using the "traditional" formula
\begin{eqnarray*}
\nu_\alpha(T)|_K
=
\beta
\|\mathbf{u}\|_{L^\infty(K)}
h_K,
\end{eqnarray*}
which we know to be stable if only \(\beta\) is large enough. Doing a couple hundred time steps (on a coarser mesh than the one shown in the program, and with a different viscosity that affects transport velocities and therefore time step sizes) in 2d will produce the following graph:
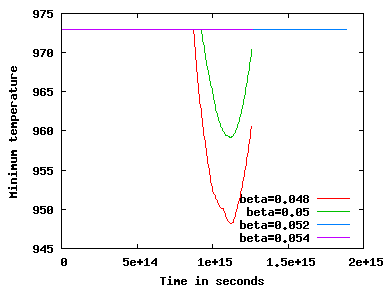
As can be seen, values \(\beta \le 0.05\) are too small whereas \(\beta=0.052\) appears to work, at least to the time horizon shown here. As a remark on the side, there are at least two questions one may wonder here: First, what happens at the time when the solution becomes unstable? Looking at the graphical output, we can see that with the unreasonably coarse mesh chosen for these experiments, around time \(t=10^{15}\) seconds the plumes of hot material that have been rising towards the cold outer boundary and have then spread sideways are starting to get close to each other, squeezing out the cold material in-between. This creates a layer of cells into which fluids flows from two opposite sides and flows out toward a third, apparently a scenario that then produce these instabilities without sufficient stabilization. Second: In step-31, we used \(\beta=0.015\cdot\text{dim}\); why does this not work here? The answer to this is not entirely clear – stabilization parameters are certainly known to depend on things like the shape of cells, for which we had squares in step-31 but have trapezoids in the current program. Whatever the exact cause, we at least have a value of \(\beta\), namely 0.052 for 2d, that works for the current program. A similar set of experiments can be made in 3d where we find that \(\beta=0.078\) is a good choice — neatly leading to the formula \(\beta=0.026 \cdot \textrm{dim}\).
With this value fixed, we can go back to the original formula for the viscosity \(\nu\) and play with the constant \(c_R\), making it as large as possible in order to make \(\nu\) as small as possible. This gives us a picture like this:

Consequently, \(c_R=0.1\) would appear to be the right value here. While this graph has been obtained for an exponent \(\alpha=1\), in the program we use \(\alpha=2\) instead, and in that case one has to re-tune the parameter (and observe that \(c_R\) appears in the numerator and not in the denominator). It turns out that \(c_R=1\) works with \(\alpha=2\).
Locally conservative Stokes discretization
The standard Taylor-Hood discretization for Stokes, using the \(Q_{k+1}^d
\times Q_k\) element, is globally conservative, i.e. \(\int_{\partial\Omega}
\mathbf n \cdot \mathbf u_h = 0\). This can easily be seen: the weak form of the divergence equation reads \((q_h, \textrm{div}\; \mathbf u_h)=0, \forall
q_h\in Q_h\). Because the pressure space does contain the function \(q_h=1\), we get
\begin{align*}
0 = (1, \textrm{div}\; \mathbf u_h)_\Omega
= \int_\Omega \textrm{div}\; \mathbf u_h
= \int_{\partial\Omega} \mathbf n \cdot \mathbf u_h
\end{align*}
by the divergence theorem. This property is important: if we want to use the velocity field \(u_h\) to transport along other quantities (such as the temperature in the current equations, but it could also be concentrations of chemical substances or entirely artificial tracer quantities) then the conservation property guarantees that the amount of the quantity advected remains constant.
That said, there are applications where this global property is not enough. Rather, we would like that it holds locally, on every cell. This can be achieved by using the space \(Q_{k+1}^d \times DGP_k\) for discretization, where we have replaced the continuous space of tensor product polynomials of degree \(k\) for the pressure by the discontinuous space of the complete polynomials of the same degree. (Note that tensor product polynomials in 2d contain the functions \(1, x, y, xy\), whereas the complete polynomials only have the functions \(1,x,y\).) This space turns out to be stable for the Stokes equation.
Because the space is discontinuous, we can now in particular choose the test function \(q_h(\mathbf x)=\chi_K(\mathbf x)\), i.e. the characteristic function of cell \(K\). We then get in a similar fashion as above
\begin{align*}
0
= (q_h, \textrm{div}\; \mathbf u_h)_\Omega
= (1, \textrm{div}\; \mathbf u_h)_K
= \int_K \textrm{div}\; \mathbf u_h
= \int_{\partial K} \mathbf n \cdot \mathbf u_h,
\end{align*}
showing the conservation property for cell \(K\). This clearly holds for each cell individually.
There are good reasons to use this discretization. As mentioned above, this element guarantees conservation of advected quantities on each cell individually. A second advantage is that the pressure mass matrix we use as a preconditioner in place of the Schur complement becomes block diagonal and consequently very easy to invert. However, there are also downsides. For one, there are now more pressure variables, increasing the overall size of the problem, although this doesn't seem to cause much harm in practice. More importantly, though, the fact that now the divergence integrated over each cell is zero when it wasn't before does not guarantee that the divergence is pointwise smaller. In fact, as one can easily verify, the \(L_2\) norm of the divergence is larger for this than for the standard Taylor-Hood discretization. (However, both converge at the same rate to zero, since it is easy to see that \(\|\textrm{div}\; u_h\|=
\|\textrm{div}\; (u-u_h)\|=
\|\textrm{trace}\; \nabla (u-u_h)\|\le
\|\nabla (u-u_h)\|={\cal O}(h^{k+2})\).) It is therefore not a priori clear that the error is indeed smaller just because we now have more degrees of freedom.
Given these considerations, it remains unclear which discretization one should prefer. Consequently, we leave that up to the user and make it a parameter in the input file which one to use.
Higher order mappings for curved boundaries
In the program, we will use a spherical shell as domain. This means that the inner and outer boundary of the domain are no longer "straight" (by which we usually mean that they are bilinear surfaces that can be represented by the FlatManifold class). Rather, they are curved and it seems prudent to use a curved approximation in the program if we are already using higher order finite elements for the velocity. Consequently, we will introduce a member variable of type MappingQ that denotes such a mapping (step-10 and step-11 introduce such mappings for the first time) and that we will use in all computations on cells that are adjacent to the boundary. Since this only affects a relatively small fraction of cells, the additional effort is not very large and we will take the luxury of using a quartic mapping for these cells.
Parallelization on clusters
Running convection codes in 3d with significant Rayleigh numbers requires a lot of computations — in the case of whole earth simulations on the order of one or several hundred million unknowns. This can obviously not be done with a single machine any more (at least not in 2010 when we started writing this code). Consequently, we need to parallelize it. Parallelization of scientific codes across multiple machines in a cluster of computers is almost always done using the Message Passing Interface (MPI). This program is no exception to that, and it follows the general spirit of the step-17 and step-18 programs in this though in practice it borrows more from step-40 in which we first introduced the classes and strategies we use when we want to completely distribute all computations, and step-55 that shows how to do that for vector-valued problems: including, for example, splitting the mesh up into a number of parts so that each processor only stores its own share plus some ghost cells, and using strategies where no processor potentially has enough memory to hold the entries of the combined solution vector locally. The goal is to run this code on hundreds or maybe even thousands of processors, at reasonable scalability.
- Note
- Even though it has a larger number, step-40 comes logically before the current program. The same is true for step-55. You will probably want to look at these programs before you try to understand what we do here.
MPI is a rather awkward interface to program with. It is a semi-object oriented set of functions, and while one uses it to send data around a network, one needs to explicitly describe the data types because the MPI functions insist on getting the address of the data as void* objects rather than deducing the data type automatically through overloading or templates. We've already seen in step-17 and step-18 how to avoid almost all of MPI by putting all the communication necessary into either the deal.II library or, in those programs, into PETSc. We'll do something similar here: like in step-40 and step-55, deal.II and the underlying p4est library are responsible for all the communication necessary for distributing the mesh, and we will let the Trilinos library (along with the wrappers in namespace TrilinosWrappers) deal with parallelizing the linear algebra components. We have already used Trilinos in step-31, and will do so again here, with the difference that we will use its parallel capabilities.
Trilinos consists of a significant number of packages, implementing basic parallel linear algebra operations (the Epetra package), different solver and preconditioner packages, and on to things that are of less importance to deal.II (e.g., optimization, uncertainty quantification, etc). deal.II's Trilinos interfaces encapsulate many of the things Trilinos offers that are of relevance to PDE solvers, and provides wrapper classes (in namespace TrilinosWrappers) that make the Trilinos matrix, vector, solver and preconditioner classes look very much the same as deal.II's own implementations of this functionality. However, as opposed to deal.II's classes, they can be used in parallel if we give them the necessary information. As a consequence, there are two Trilinos classes that we have to deal with directly (rather than through wrappers), both of which are part of Trilinos' Epetra library of basic linear algebra and tool classes:
-
The Epetra_Comm class is an abstraction of an MPI "communicator", i.e., it describes how many and which machines can communicate with each other. Each distributed object, such as a sparse matrix or a vector for which we may want to store parts on different machines, needs to have a communicator object to know how many parts there are, where they can be found, and how they can be accessed.
In this program, we only really use one communicator object – based on the MPI variable MPI_COMM_WORLD – that encompasses all processes that work together. It would be perfectly legitimate to start a process on \(N\) machines but only store vectors on a subset of these by producing a communicator object that only encompasses this subset of machines; there is really no compelling reason to do so here, however.
-
The IndexSet class is used to describe which elements of a vector or which rows of a matrix should reside on the current machine that is part of a communicator. To create such an object, you need to know (i) the total number of elements or rows, (ii) the indices of the elements you want to store locally. We will set up these partitioners in the BoussinesqFlowProblem::setup_dofs function below and then hand it to every parallel object we create.
Unlike PETSc, Trilinos makes no assumption that the elements of a vector need to be partitioned into contiguous chunks. At least in principle, we could store all elements with even indices on one processor and all odd ones on another. That's not very efficient, of course, but it's possible. Furthermore, the elements of these partitionings do not necessarily be mutually exclusive. This is important because when postprocessing solutions, we need access to all locally relevant or at least the locally active degrees of freedom (see the module on Parallel computing with multiple processors using distributed memory for a definition, as well as the discussion in step-40). Which elements the Trilinos vector considers as locally owned is not important to us then. All we care about is that it stores those elements locally that we need.
There are a number of other concepts relevant to distributing the mesh to a number of processors; you may want to take a look at the Parallel computing with multiple processors using distributed memory module and step-40 or step-55 before trying to understand this program. The rest of the program is almost completely agnostic about the fact that we don't store all objects completely locally. There will be a few points where we have to limit loops over all cells to those that are locally owned, or where we need to distinguish between vectors that store only locally owned elements and those that store everything that is locally relevant (see this glossary entry), but by and large the amount of heavy lifting necessary to make the program run in parallel is well hidden in the libraries upon which this program builds. In any case, we will comment on these locations as we get to them in the program code.
Parallelization within individual nodes of a cluster
The second strategy to parallelize a program is to make use of the fact that most computers today have more than one processor that all have access to the same memory. In other words, in this model, we don't explicitly have to say which pieces of data reside where – all of the data we need is directly accessible and all we have to do is split processing this data between the available processors. We will then couple this with the MPI parallelization outlined above, i.e., we will have all the processors on a machine work together to, for example, assemble the local contributions to the global matrix for the cells that this machine actually "owns" but not for those cells that are owned by other machines. We will use this strategy for four kinds of operations we frequently do in this program: assembly of the Stokes and temperature matrices, assembly of the matrix that forms the Stokes preconditioner, and assembly of the right hand side of the temperature system.
All of these operations essentially look as follows: we need to loop over all cells for which cell->subdomain_id() equals the index our machine has within the communicator object used for all communication (i.e., MPI_COMM_WORLD, as explained above). The test we are actually going to use for this, and which describes in a concise way why we test this condition, is cell->is_locally_owned(). On each such cell we need to assemble the local contributions to the global matrix or vector, and then we have to copy each cell's contribution into the global matrix or vector. Note that the first part of this (the loop) defines a range of iterators on which something has to happen. The second part, assembly of local contributions is something that takes the majority of CPU time in this sequence of steps, and is a typical example of things that can be done in parallel: each cell's contribution is entirely independent of all other cells' contributions. The third part, copying into the global matrix, must not happen in parallel since we are modifying one object and so several threads can not at the same time read an existing matrix element, add their contribution, and write the sum back into memory without danger of producing a race condition.
deal.II has a class that is made for exactly this workflow: WorkStream, first discussed in step-9 and step-13. Its use is also extensively documented in the module on Parallel computing with multiple processors accessing shared memory (in the section on the WorkStream class) and we won't repeat here the rationale and detailed instructions laid out there, though you will want to read through this module to understand the distinction between scratch space and per-cell data. Suffice it to mention that we need the following:
- An iterator range for those cells on which we are supposed to work. This is provided by the FilteredIterator class which acts just like every other cell iterator in deal.II with the exception that it skips all cells that do not satisfy a particular predicate (i.e., a criterion that evaluates to true or false). In our case, the predicate is whether a cell is locally owned.
- A function that does the work on each cell for each of the tasks identified above, i.e., functions that assemble the local contributions to Stokes matrix and preconditioner, temperature matrix, and temperature right hand side. These are the
BoussinesqFlowProblem::local_assemble_stokes_system, BoussinesqFlowProblem::local_assemble_stokes_preconditioner, BoussinesqFlowProblem::local_assemble_temperature_matrix, and BoussinesqFlowProblem::local_assemble_temperature_rhs functions in the code below. These four functions can all have several instances running in parallel at the same time.
- Functions that copy the result of the previous ones into the global object and that run sequentially to avoid race conditions. These are the
BoussinesqFlowProblem::copy_local_to_global_stokes_system, BoussinesqFlowProblem::copy_local_to_global_stokes_preconditioner, BoussinesqFlowProblem::copy_local_to_global_temperature_matrix, and BoussinesqFlowProblem::copy_local_to_global_temperature_rhs functions.
We will comment on a few more points in the actual code, but in general their structure should be clear from the discussion in Parallel computing with multiple processors accessing shared memory.
The underlying technology for WorkStream identifies "tasks" that need to be worked on (e.g. assembling local contributions on a cell) and schedules these tasks automatically to available processors. WorkStream creates these tasks automatically, by splitting the iterator range into suitable chunks.
- Note
- Using multiple threads within each MPI process only makes sense if you have fewer MPI processes running on each node of your cluster than there are processor cores on this machine. Otherwise, MPI will already keep your processors busy and you won't get any additional speedup from using threads. For example, if your cluster nodes have 8 cores as they often have at the time of writing this, and if your batch scheduler puts 8 MPI processes on each node, then using threads doesn't make the program any faster. Consequently, you probably want to either configure your deal.II without threads, or set the number of threads in Utilities::MPI::MPI_InitFinalize to 1 (third argument), or "export DEAL_II_NUM_THREADS=1" before running. That said, at the time of writing this, we only use the WorkStream class for assembling (parts of) linear systems, while 75% or more of the run time of the program is spent in the linear solvers that are not parallelized — in other words, the best we could hope is to parallelize the remaining 25%.
The testcase
The setup for this program is mildly reminiscent of the problem we wanted to solve in the first place (see the introduction of step-31): convection in the earth mantle. As a consequence, we choose the following data, all of which appears in the program in units of meters and seconds (the SI system) even if we list them here in other units. We do note, however, that these choices are essentially still only exemplary, and not meant to result in a completely realistic description of convection in the earth mantle: for that, more and more difficult physics would have to be implemented, and several other aspects are currently missing from this program as well. We will come back to this issue in the results section again, but state for now that providing a realistic description is a goal of the ASPECT code in development at the time of writing this.
As a reminder, let us again state the equations we want to solve are these:
\begin{eqnarray*}
-\nabla \cdot (2 \eta \varepsilon ({\mathbf u})) +
\nabla \left( \frac{\eta}{L} \hat p\right) &=&
\rho(T) \mathbf{g},
\\
\frac{\eta}{L} \nabla \cdot {\mathbf u} &=& 0,
\\
\frac{\partial T}{\partial t}
+
{\mathbf u} \cdot \nabla T
-
\nabla \cdot \kappa \nabla T &=& \gamma,
\end{eqnarray*}
augmented by boundary and initial conditions. We then have to choose data for the following quantities:
-
The domain is an annulus (in 2d) or a spherical shell (in 3d) with inner and outer radii that match that of the earth: the total radius of the earth is 6371km, with the mantle starting at a depth of around 35km (just under the solid earth crust composed of continental and oceanic plates) to a depth of 2890km (where the outer earth core starts). The radii are therefore \(R_0=(6371-2890)\text{km},
R_1=(6371-35)\text{km}\). This domain is conveniently generated using the GridGenerator::hyper_shell() function.
-
At the interface between crust and mantle, the temperature is between 500 and 900 degrees Celsius, whereas at its bottom it is around 4000 degrees Celsius (see, for example, this Wikipedia entry). In Kelvin, we therefore choose \(T_0=(4000+273)\text{K}\), \(T_1=(500+273)\text{K}\) as boundary conditions at the inner and outer edge.
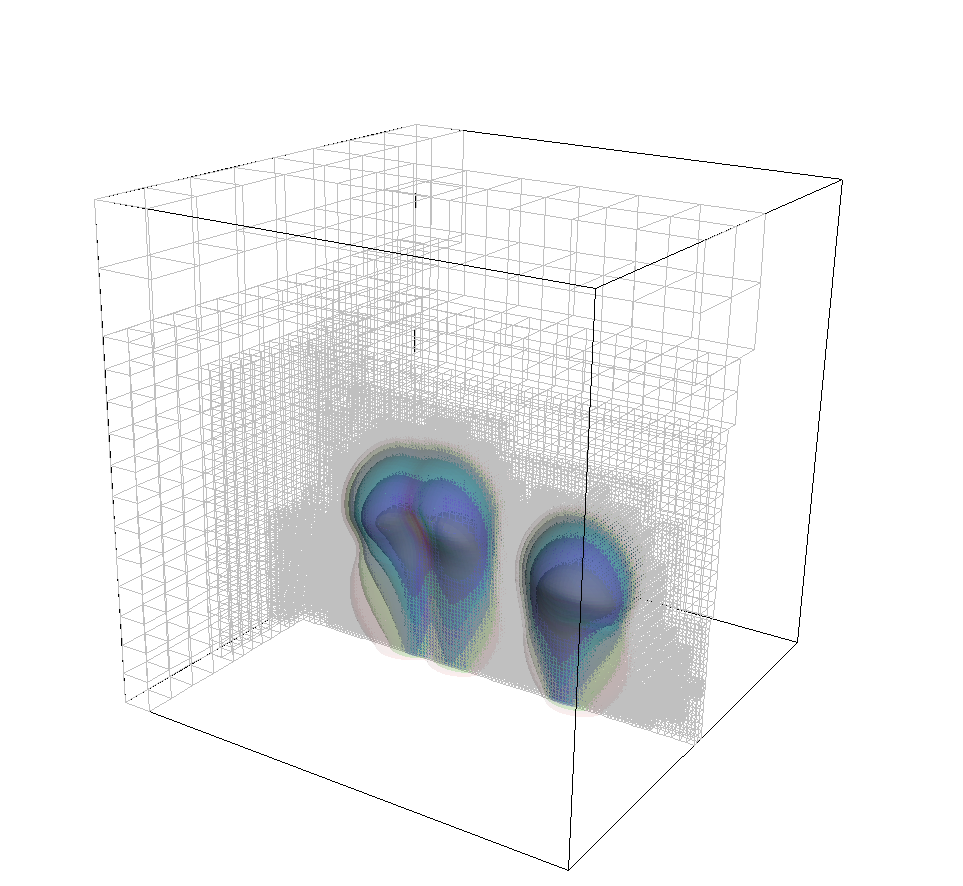
In addition to this, we also have to specify some initial conditions for the temperature field. The real temperature field of the earth is quite complicated as a consequence of the convection that has been going on for more than four billion years – in fact, it is the properties of this temperature distribution that we want to explore with programs like this. As a consequence, we don't really have anything useful to offer here, but we can hope that if we start with something and let things run for a while that the exact initial conditions don't matter that much any more — as is in fact suggested by looking at the pictures shown in the results section below. The initial temperature field we use here is given in terms of the radius by
\begin{align*}
s &= \frac{\|\mathbf x\|-R_0}{R_1-R_0}, \\
\varphi &= \arctan \frac{y}{x}, \\
\tau &= s + \frac 15 s(1-s) \sin(6\varphi) q(z), \\
T(\mathbf x) &= T_0(1-\tau) + T_1\tau,
\end{align*}
where
\begin{align*}
q(z) = \left\{
\begin{array}{ll}
1 & \text{in 2d} \\
\max\{0, \cos(\pi |z/R_1|)\} & \text{in 3d}
\end{array}
\right. .
\end{align*}
This complicated function is essentially a perturbation of a linear profile between the inner and outer temperatures. In 2d, the function \(\tau=\tau(\mathbf x)\) looks like this (I got the picture from this page):
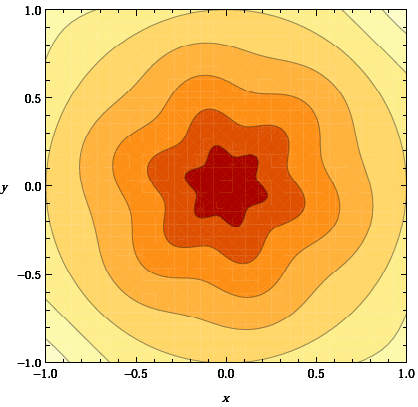
The point of this profile is that if we had used \(s\) instead of \(\tau\) in the definition of \(T(\mathbf x)\) then it would simply be a linear interpolation. \(\tau\) has the same function values as \(s\) on the inner and outer boundaries (zero and one, respectively), but it stretches the temperature profile a bit depending on the angle and the \(z\) value in 3d, producing an angle-dependent perturbation of the linearly interpolating field. We will see in the results section that this is an entirely unphysical temperature field (though it will make for interesting images) as the equilibrium state for the temperature will be an almost constant temperature with boundary layers at the inner and outer boundary.
-
The right hand side of the temperature equation contains the rate of internal heating \(\gamma\). The earth does heat naturally through several mechanisms: radioactive decay, chemical separation (heavier elements sink to the bottom, lighter ones rise to the top; the countercurrents dissipate energy equal to the loss of potential energy by this separation process); heat release by crystallization of liquid metal as the solid inner core of the earth grows; and heat dissipation from viscous friction as the fluid moves.
Chemical separation is difficult to model since it requires modeling mantle material as multiple phases; it is also a relatively small effect. Crystallization heat is even more difficult since it is confined to areas where temperature and pressure allow for phase changes, i.e., a discontinuous process. Given the difficulties in modeling these two phenomena, we will neglect them.
The other two are readily handled and, given the way we scaled the temperature equation, lead to the equation
\[
\gamma(\mathbf x)
=
\frac{\rho q+2\eta \varepsilon(\mathbf u):\varepsilon(\mathbf u)}
{\rho c_p},
\]
where \(q\) is the radiogenic heating in \(\frac{W}{kg}\), and the second term in the enumerator is viscous friction heating. \(\rho\) is the density and \(c_p\) is the specific heat. The literature provides the following approximate values: \(c_p=1250 \frac{J}{kg\; K}, q=7.4\cdot 10^{-12}\frac{W}{kg}\). The other parameters are discussed elsewhere in this section.
We neglect one internal heat source, namely adiabatic heating here, which will lead to a surprising temperature field. This point is commented on in detail in the results section below.
-
For the velocity we choose as boundary conditions \(\mathbf{v}=0\) at the inner radius (i.e., the fluid sticks to the earth core) and \(\mathbf{n}\cdot\mathbf{v}=0\) at the outer radius (i.e., the fluid flows tangentially along the bottom of the earth crust). Neither of these is physically overly correct: certainly, on both boundaries, fluids can flow tangentially, but they will incur a shear stress through friction against the medium at the other side of the interface (the metallic core and the crust, respectively). Such a situation could be modeled by a Robin-type boundary condition for the tangential velocity; in either case, the normal (vertical) velocity would be zero, although even that is not entirely correct since continental plates also have vertical motion (see, for example, the phenomenon of post-glacial rebound). But to already make things worse for the tangential velocity, the medium on the other side is in motion as well, so the shear stress would, in the simplest case, be proportional to the velocity difference, leading to a boundary condition of the form
\begin{align*}
\mathbf{n}\cdot [2\eta \varepsilon(\mathbf v)]
&=
s \mathbf{n} \times [\mathbf v - \mathbf v_0],
\\
\mathbf{n} \cdot \mathbf v &= 0,
\end{align*}
with a proportionality constant \(s\). Rather than going down this route, however, we go with the choice of zero (stick) and tangential flow boundary conditions.
As a side note of interest, we may also have chosen tangential flow conditions on both inner and outer boundary. That has a significant drawback, however: it leaves the velocity not uniquely defined. The reason is that all velocity fields \(\hat{\mathbf v}\) that correspond to a solid body rotation around the center of the domain satisfy \(\mathrm{div}\;
\varepsilon(\hat{\mathbf v})=0, \mathrm{div} \;\hat{\mathbf v} = 0\), and \(\mathbf{n} \cdot \hat{\mathbf v} = 0\). As a consequence, if \(\mathbf v\) satisfies equations and boundary conditions, then so does \(\mathbf v +
\hat{\mathbf v}\). That's certainly not a good situation that we would like to avoid. The traditional way to work around this is to pick an arbitrary point on the boundary and call this your fixed point by choosing the velocity to be zero in all components there. (In 3d one has to choose two points.) Since this program isn't meant to be too realistic to begin with, we avoid this complication by simply fixing the velocity along the entire interior boundary.
-
To first order, the gravity vector always points downward. The question for a body as big as the earth is just: where is "up". The naive answer of course is "radially inward, towards the center of the earth". So at the surface of the earth, we have
\[
\mathbf g
=
-9.81 \frac{\text{m}}{\text{s}^2} \frac{\mathbf x}{\|\mathbf x\|},
\]
where \(9.81 \frac{\text{m}}{\text{s}^2}\) happens to be the average gravity acceleration at the earth surface. But in the earth interior, the question becomes a bit more complicated: at the (bary-)center of the earth, for example, you have matter pulling equally hard in all directions, and so \(\mathbf g=0\). In between, the net force is described as follows: let us define the gravity potential by
\[
\varphi(\mathbf x)
=
\int_{\text{earth}}
-G \frac{\rho(\mathbf y)}{\|\mathbf x-\mathbf y\|}
\ \text{d}y,
\]
then \(\mathbf g(\mathbf x) = -\nabla \varphi(\mathbf x)\). If we assume that the density \(\rho\) is constant throughout the earth, we can produce an analytical expression for the gravity vector (don't try to integrate above equation somehow – it leads to elliptic integrals; a simpler way is to notice that \(-\Delta\varphi(\mathbf x) = -4\pi G \rho
\chi_{\text{earth}}(\mathbf x)\) and solving this partial differential equation in all of \({\mathbb R}^3\) exploiting the radial symmetry):
\[
\mathbf g(\mathbf x) =
\left\{
\begin{array}{ll}
-\frac{4}{3}\pi G \rho \|\mathbf x\| \frac{\mathbf x}{\|\mathbf x\|}
& \text{for} \ \|\mathbf x\|<R_1, \\
-\frac{4}{3}\pi G \rho R^3 \frac{1}{\|\mathbf x\|^2}
\frac{\mathbf x}{\|\mathbf x\|}
& \text{for} \ \|\mathbf x\|\ge R_1.
\end{array}
\right.
\]
The factor \(-\frac{\mathbf x}{\|\mathbf x\|}\) is the unit vector pointing radially inward. Of course, within this problem, we are only interested in the branch that pertains to within the earth, i.e., \(\|\mathbf
x\|<R_1\). We would therefore only consider the expression
\[
\mathbf g(\mathbf x) =
-\frac{4}{3}\pi G \rho \|\mathbf x\| \frac{\mathbf x}{\|\mathbf x\|}
=
-\frac{4}{3}\pi G \rho \mathbf x
=
- 9.81 \frac{\mathbf x}{R_1} \frac{\text{m}}{\text{s}^2},
\]
where we can infer the last expression because we know Earth's gravity at the surface (where \(\|x\|=R_1\)).
One can derive a more general expression by integrating the differential equation for \(\varphi(r)\) in the case that the density distribution is radially symmetric, i.e., \(\rho(\mathbf
x)=\rho(\|\mathbf x\|)=\rho(r)\). In that case, one would get
\[
\varphi(r)
= 4\pi G \int_0^r \frac 1{s^2} \int_0^s t^2 \rho(t) \; dt \; ds.
\]
There are two problems with this, however: (i) The Earth is not homogeneous, i.e., the density \(\rho\) depends on \(\mathbf x\); in fact it is not even a function that only depends on the radius \(r=\|\mathbf x\|\). In reality, gravity therefore does not always decrease as we get deeper: because the earth core is so much denser than the mantle, gravity actually peaks at around \(10.7
\frac{\text{m}}{\text{s}^2}\) at the core mantle boundary (see this article). (ii) The density, and by consequence the gravity vector, is not even constant in time: after all, the problem we want to solve is the time dependent upwelling of hot, less dense material and the downwelling of cold dense material. This leads to a gravity vector that varies with space and time, and does not always point straight down.
In order to not make the situation more complicated than necessary, we could use the approximation that at the inner boundary of the mantle, gravity is \(10.7 \frac{\text{m}}{\text{s}^2}\) and at the outer boundary it is \(9.81 \frac{\text{m}}{\text{s}^2}\), in each case pointing radially inward, and that in between gravity varies linearly with the radial distance from the earth center. That said, it isn't that hard to actually be slightly more realistic and assume (as we do below) that the earth mantle has constant density. In that case, the equation above can be integrated and we get an expression for \(\|\mathbf{g}\|\) where we can fit constants to match the gravity at the top and bottom of the earth mantle to obtain
\[
\|\mathbf{g}\|
= 1.245\cdot 10^{-6} \frac{1}{\textrm{s}^2} r + 7.714\cdot 10^{13} \frac{\textrm{m}^3}{\textrm{s}^2}\frac{1}{r^2}.
\]
-
The density of the earth mantle varies spatially, but not by very much. \(\rho_{\text{ref}}=3300 \frac{\text{kg}}{\text{m}^3}\) is a relatively good average value for the density at reference temperature \(T_{\text{ref}}=293\) Kelvin.
-
The thermal expansion coefficient \(\beta\) also varies with depth (through its dependence on temperature and pressure). Close to the surface, it appears to be on the order of \(\beta=45\cdot 10^{-6} \frac 1{\text{K}}\), whereas at the core mantle boundary, it may be closer to \(\beta=10\cdot
10^{-6} \frac 1{\text{K}}\). As a reasonable value, let us choose \(\beta=2\cdot 10^{-5} \frac 1{\text{K}}\). The density as a function of temperature is then \(\rho(T)=[1-\beta(T-T_{\text{ref}})]\rho_{\text{ref}}\).
-
The second to last parameter we need to specify is the viscosity \(\eta\). This is a tough one, because rocks at the temperatures and pressure typical for the earth mantle flow so slowly that the viscosity can not be determined accurately in the laboratory. So how do we know about the viscosity of the mantle? The most commonly used route is to consider that during and after ice ages, ice shields form and disappear on time scales that are shorter than the time scale of flow in the mantle. As a consequence, continents slowly sink into the earth mantle under the added weight of an ice shield, and they rise up again slowly after the ice shield has disappeared again (this is called postglacial rebound). By measuring the speed of this rebound, we can infer the viscosity of the material that flows into the area vacated under the rebounding continental plates.
Using this technique, values around \(\eta=10^{21} \text{Pa}\;\text{s}
= 10^{21} \frac{\text{N}\;\text{s}}{\text{m}^2}
= 10^{21} \frac{\text{kg}}{\text{m}\;\text{s}}\) have been found as the most likely, though the error bar on this is at least one order of magnitude.
While we will use this value, we again have to caution that there are many physical reasons to assume that this is not the correct value. First, it should really be made dependent on temperature: hotter material is most likely to be less viscous than colder material. In reality, however, the situation is even more complex. Most rocks in the mantle undergo phase changes as temperature and pressure change: depending on temperature and pressure, different crystal configurations are thermodynamically favored over others, even if the chemical composition of the mantle were homogeneous. For example, the common mantle material MgSiO3 exists in its perovskite structure throughout most of the mantle, but in the lower mantle the same substance is stable only as post-perovskite. Clearly, to compute realistic viscosities, we would not only need to know the exact chemical composition of the mantle and the viscosities of all materials, but we would also have to compute the thermodynamically most stable configurations for all materials at each quadrature point. This is at the time of writing this program not a feasible suggestion.
-
Our last material parameter is the thermal diffusivity \(\kappa\), which is defined as \(\kappa=\frac{k}{\rho c_p}\) where \(k\) is the thermal conductivity, \(\rho\) the density, and \(c_p\) the specific heat. For this, the literature indicates that it increases from around \(0.7\) in the upper mantle to around \(1.7 \frac{\text{mm}^2}{\text{s}}\) in the lower mantle, though the exact value is not really all that important: heat transport through convection is several orders of magnitude more important than through thermal conduction. It may be of interest to know that perovskite, the most abundant material in the earth mantle, appears to become transparent at pressures above around 120 GPa (see, for example, J. Badro et al., Science 305, 383-386 (2004)); in the lower mantle, it may therefore be that heat transport through radiative transfer is more efficient than through thermal conduction.
In view of these considerations, let us choose \(\kappa=1 \frac{\text{mm}^2}{\text{s}} =10^{-6} \frac{\text{m}^2}{\text{s}}\) for the purpose of this program.
All of these pieces of equation data are defined in the program in the EquationData namespace. When run, the program produces long-term maximal velocities around 10-40 centimeters per year (see the results section below), approximately the physically correct order of magnitude. We will set the end time to 1 billion years.
- Note
- The choice of the constants and material parameters above follows in large part the comprehensive book "Mantle Convection in the Earth and Planets,
Part 1" by G. Schubert and D. L. Turcotte and P. Olson (Cambridge, 2001). It contains extensive discussion of ways to make the program more realistic.
Implementation details
Compared to step-31, this program has a number of noteworthy differences:
- The
EquationData namespace is significantly larger, reflecting the fact that we now have much more physics to deal with. That said, most of this additional physical detail is rather self-contained in functions in this one namespace, and does not proliferate throughout the rest of the program.
- Of more obvious visibility is the fact that we have put a good number of parameters into an input file handled by the ParameterHandler class (see, for example, step-29, for ways to set up run-time parameter files with this class). This often makes sense when one wants to avoid re-compiling the program just because one wants to play with a single parameter (think, for example, of parameter studies determining the best values of the stabilization constants discussed above), in particular given that it takes a nontrivial amount of time to re-compile programs of the current size. To just give an overview of the kinds of parameters we have moved from fixed values into the input file, here is a listing of a typical
step-32.prm file : # Listing of Parameters
# ---------------------
# The end time of the simulation in years.
# Whether graphical output is to be generated or not. You may not want to get
# graphical output if the number of processors is large.
set Generate graphical output =
false
# The number of adaptive refinement steps performed after initial global
# refinement.
set Initial adaptive refinement = 1
# The number of global refinement steps performed on the initial coarse mesh,
# before the problem is first solved there.
set Initial global refinement = 1
# The number of time steps between each generation of graphical output files.
set Time steps between graphical output = 50
# The number of time steps after which the mesh is to be adapted based on
# computed error indicators.
set Time steps between mesh refinement = 10
subsection Discretization
# The polynomial degree to use for the velocity variables in the Stokes
# system.
set Stokes velocity polynomial degree = 2
# The polynomial degree to use for the temperature variable.
set Temperature polynomial degree = 2
# Whether to use a Stokes discretization that is locally conservative at the
# expense of a larger number of degrees of freedom, or to go with a cheaper
# discretization that does not locally conserve mass (although it is
# globally conservative.
set Use locally conservative discretization =
true
subsection Stabilization parameters
# The exponent in the entropy viscosity stabilization.
# The beta factor in the artificial viscosity stabilization. An appropriate
# value for 2d is 0.052 and 0.078 for 3d.
# The c_R factor in the entropy viscosity stabilization.
__global__ void set(Number *val, const Number s, const size_type N)
VectorType::value_type * end(VectorType &V)
- There are, obviously, a good number of changes that have to do with the fact that we want to run our program on a possibly very large number of machines. Although one may suspect that this requires us to completely re-structure our code, that isn't in fact the case (although the classes that implement much of this functionality in deal.II certainly look very different from an implementation viewpoint, but this doesn't reflect in their public interface). Rather, the changes are mostly subtle, and the overall structure of the main class is pretty much unchanged. That said, the devil is in the detail: getting parallel computing right, without deadlocks, ensuring that the right data is available at the right place (see, for example, the discussion on fully distributed vectors vs. vectors with ghost elements), and avoiding bottlenecks is difficult and discussions on this topic will appear in a good number of places in this program.
Outlook
This is a tutorial program. That means that at least most of its focus needs to lie on demonstrating ways of using deal.II and associated libraries, and not diluting this teaching lesson by focusing overly much on physical details. Despite the lengthy section above on the choice of physical parameters, the part of the program devoted to this is actually quite short and self contained.
That said, both step-31 and the current step-32 have not come about by chance but are certainly meant as wayposts along the path to a more comprehensive program that will simulate convection in the earth mantle. We call this code ASPECT (short for Advanced Solver for Problems in Earth's ConvecTion); its development is funded by the Computational Infrastructure in Geodynamics initiative with support from the National Science Foundation. More information on ASPECT is available at its homepage.
The commented program
Include files
The first task as usual is to include the functionality of these well-known deal.II library files and some C++ header files.
#include <fstream>
#include <iostream>
#include <limits>
#include <locale>
#include <string>
This is the only include file that is new: It introduces the parallel::distributed::SolutionTransfer equivalent of the SolutionTransfer class to take a solution from on mesh to the next one upon mesh refinement, but in the case of parallel distributed triangulations:
The following classes are used in parallel distributed computations and have all already been introduced in step-40:
The next step is like in all previous tutorial programs: We put everything into a namespace of its own and then import the deal.II classes and functions into it:
Equation data
In the following namespace, we define the various pieces of equation data that describe the problem. This corresponds to the various aspects of making the problem at least slightly realistic and that were exhaustively discussed in the description of the testcase in the introduction.
We start with a few coefficients that have constant values (the comment after the value indicates its physical units):
namespace EquationData
{
constexpr double eta = 1e21;
constexpr double kappa = 1
e-6;
constexpr double reference_density = 3300;
constexpr double reference_temperature = 293;
constexpr double expansion_coefficient = 2
e-5;
constexpr double specific_heat = 1250;
constexpr double radiogenic_heating = 7.4e-12;
constexpr double R0 = 6371000. - 2890000.;
constexpr double R1 = 6371000. - 35000.;
constexpr double T0 = 4000 + 273;
constexpr double T1 = 700 + 273;
SymmetricTensor< 2, dim, Number > e(const Tensor< 2, dim, Number > &F)
The next set of definitions are for functions that encode the density as a function of temperature, the gravity vector, and the initial values for the temperature. Again, all of these (along with the values they compute) are discussed in the introduction:
double density(const double temperature)
{
return (
reference_density *
(1 - expansion_coefficient * (temperature - reference_temperature)));
}
template <int dim>
{
const double r = p.
norm();
return -(1.245e-6 * r + 7.714e13 / r / r) * p / r;
}
template <int dim>
class TemperatureInitialValues :
public Function<dim>
{
public:
TemperatureInitialValues()
{}
const unsigned int component = 0) const override;
};
template <int dim>
double TemperatureInitialValues<dim>::value(
const Point<dim> &p,
const unsigned int) const
{
const double r = p.
norm();
const double h = R1 - R0;
const double s = (r - R0) / h;
const double q =
const double tau = s + 0.2 * s * (1 - s) *
std::sin(6 * phi) * q;
return T0 * (1.0 - tau) + T1 * tau;
}
template <int dim>
void
TemperatureInitialValues<dim>::vector_value(
const Point<dim> &p,
{
for (unsigned int c = 0; c < this->n_components; ++c)
values(c) = TemperatureInitialValues<dim>::value(p, c);
}
virtual RangeNumberType value(const Point< dim > &p, const unsigned int component=0) const
virtual void vector_value(const Point< dim > &p, Vector< RangeNumberType > &values) const
numbers::NumberTraits< Number >::real_type norm() const
VectorizedArray< Number, width > abs(const ::VectorizedArray< Number, width > &x)
VectorizedArray< Number, width > cos(const ::VectorizedArray< Number, width > &x)
Expression atan2(const Expression &y, const Expression &x)
static constexpr double PI
::VectorizedArray< Number, width > max(const ::VectorizedArray< Number, width > &, const ::VectorizedArray< Number, width > &)
::VectorizedArray< Number, width > sin(const ::VectorizedArray< Number, width > &)
As mentioned in the introduction we need to rescale the pressure to avoid the relative ill-conditioning of the momentum and mass conservation equations. The scaling factor is \(\frac{\eta}{L}\) where \(L\) was a typical length scale. By experimenting it turns out that a good length scale is the diameter of plumes, which is around 10 km:
constexpr double pressure_scaling = eta / 10000;
The final number in this namespace is a constant that denotes the number of seconds per (average, tropical) year. We use this only when generating screen output: internally, all computations of this program happen in SI units (kilogram, meter, seconds) but writing geological times in seconds yields numbers that one can't relate to reality, and so we convert to years using the factor defined here:
const double year_in_seconds = 60 * 60 * 24 * 365.2425;
}
Preconditioning the Stokes system
This namespace implements the preconditioner. As discussed in the introduction, this preconditioner differs in a number of key portions from the one used in step-31. Specifically, it is a right preconditioner, implementing the matrix
\begin{align*}
\left(\begin{array}{cc}A^{-1} & B^T
\\0 & S^{-1}
\end{array}\right)
\end{align*}
where the two inverse matrix operations are approximated by linear solvers or, if the right flag is given to the constructor of this class, by a single AMG V-cycle for the velocity block. The three code blocks of the vmult function implement the multiplications with the three blocks of this preconditioner matrix and should be self explanatory if you have read through step-31 or the discussion of composing solvers in step-20.
namespace LinearSolvers
{
template <class PreconditionerTypeA, class PreconditionerTypeMp>
{
public:
const PreconditionerTypeMp &Mppreconditioner,
const PreconditionerTypeA & Apreconditioner,
const bool do_solve_A)
: stokes_matrix(&S)
, stokes_preconditioner_matrix(&Spre)
, mp_preconditioner(Mppreconditioner)
, a_preconditioner(Apreconditioner)
, do_solve_A(do_solve_A)
{}
{
{
solver.solve(stokes_preconditioner_matrix->block(1, 1),
mp_preconditioner);
}
{
stokes_matrix->block(0, 1).vmult(utmp, dst.
block(1));
utmp *= -1.0;
}
if (do_solve_A == true)
{
solver.solve(stokes_matrix->block(0, 0),
utmp,
a_preconditioner);
}
else
a_preconditioner.vmult(dst.
block(0), utmp);
}
private:
stokes_matrix;
stokes_preconditioner_matrix;
const PreconditionerTypeMp &mp_preconditioner;
const PreconditionerTypeA & a_preconditioner;
const bool do_solve_A;
};
}
BlockType & block(const unsigned int i)
Definition of assembly data structures
As described in the introduction, we will use the WorkStream mechanism discussed in the Parallel computing with multiple processors accessing shared memory module to parallelize operations among the processors of a single machine. The WorkStream class requires that data is passed around in two kinds of data structures, one for scratch data and one to pass data from the assembly function to the function that copies local contributions into global objects.
The following namespace (and the two sub-namespaces) contains a collection of data structures that serve this purpose, one pair for each of the four operations discussed in the introduction that we will want to parallelize. Each assembly routine gets two sets of data: a Scratch array that collects all the classes and arrays that are used for the calculation of the cell contribution, and a CopyData array that keeps local matrices and vectors which will be written into the global matrix. Whereas CopyData is a container for the final data that is written into the global matrices and vector (and, thus, absolutely necessary), the Scratch arrays are merely there for performance reasons — it would be much more expensive to set up a FEValues object on each cell, than creating it only once and updating some derivative data.
step-31 had four assembly routines: One for the preconditioner matrix of the Stokes system, one for the Stokes matrix and right hand side, one for the temperature matrices and one for the right hand side of the temperature equation. We here organize the scratch arrays and CopyData objects for each of those four assembly components using a struct environment (since we consider these as temporary objects we pass around, rather than classes that implement functionality of their own, though this is a more subjective point of view to distinguish between structs and classes).
Regarding the Scratch objects, each struct is equipped with a constructor that creates an FEValues object using the FiniteElement, Quadrature, Mapping (which describes the interpolation of curved boundaries), and The interplay of UpdateFlags, Mapping, and FiniteElement in FEValues instances. Moreover, we manually implement a copy constructor (since the FEValues class is not copyable by itself), and provide some additional vector fields that are used to hold intermediate data during the computation of local contributions.
Let us start with the scratch arrays and, specifically, the one used for assembly of the Stokes preconditioner:
namespace Assembly
{
namespace Scratch
{
template <int dim>
struct StokesPreconditioner
{
StokesPreconditioner(const StokesPreconditioner &data);
std::vector<Tensor<2, dim>> grad_phi_u;
std::vector<double> phi_p;
};
template <int dim>
StokesPreconditioner<dim>::StokesPreconditioner(
: stokes_fe_values(mapping, stokes_fe, stokes_quadrature, update_flags)
, grad_phi_u(stokes_fe.n_dofs_per_cell())
, phi_p(stokes_fe.n_dofs_per_cell())
{}
template <int dim>
StokesPreconditioner<dim>::StokesPreconditioner(
const StokesPreconditioner &scratch)
: stokes_fe_values(scratch.stokes_fe_values.get_mapping(),
scratch.stokes_fe_values.get_fe(),
scratch.stokes_fe_values.get_quadrature(),
scratch.stokes_fe_values.get_update_flags())
, grad_phi_u(scratch.grad_phi_u)
, phi_p(scratch.phi_p)
{}
Abstract base class for mapping classes.
The next one is the scratch object used for the assembly of the full Stokes system. Observe that we derive the StokesSystem scratch class from the StokesPreconditioner class above. We do this because all the objects that are necessary for the assembly of the preconditioner are also needed for the actual matrix system and right hand side, plus some extra data. This makes the program more compact. Note also that the assembly of the Stokes system and the temperature right hand side further down requires data from temperature and velocity, respectively, so we actually need two FEValues objects for those two cases.
template <int dim>
struct StokesSystem : public StokesPreconditioner<dim>
{
StokesSystem(const StokesSystem<dim> &data);
std::vector<Tensor<1, dim>> phi_u;
std::vector<SymmetricTensor<2, dim>> grads_phi_u;
std::vector<double> div_phi_u;
std::vector<double> old_temperature_values;
};
template <int dim>
StokesSystem<dim>::StokesSystem(
: StokesPreconditioner<dim>(stokes_fe,
stokes_quadrature,
mapping,
stokes_update_flags)
, temperature_fe_values(mapping,
temperature_fe,
stokes_quadrature,
temperature_update_flags)
, phi_u(stokes_fe.n_dofs_per_cell())
, grads_phi_u(stokes_fe.n_dofs_per_cell())
, div_phi_u(stokes_fe.n_dofs_per_cell())
, old_temperature_values(stokes_quadrature.size())
{}
template <int dim>
StokesSystem<dim>::StokesSystem(const StokesSystem<dim> &scratch)
: StokesPreconditioner<dim>(scratch)
, temperature_fe_values(
scratch.temperature_fe_values.get_mapping(),
scratch.temperature_fe_values.get_fe(),
scratch.temperature_fe_values.get_quadrature(),
scratch.temperature_fe_values.get_update_flags())
, phi_u(scratch.phi_u)
, grads_phi_u(scratch.grads_phi_u)
, div_phi_u(scratch.div_phi_u)
, old_temperature_values(scratch.old_temperature_values)
{}
After defining the objects used in the assembly of the Stokes system, we do the same for the assembly of the matrices necessary for the temperature system. The general structure is very similar:
template <int dim>
struct TemperatureMatrix
{
TemperatureMatrix(const TemperatureMatrix &data);
std::vector<double> phi_T;
std::vector<Tensor<1, dim>> grad_phi_T;
};
template <int dim>
TemperatureMatrix<dim>::TemperatureMatrix(
: temperature_fe_values(mapping,
temperature_fe,
temperature_quadrature,
, phi_T(temperature_fe.n_dofs_per_cell())
, grad_phi_T(temperature_fe.n_dofs_per_cell())
{}
template <int dim>
TemperatureMatrix<dim>::TemperatureMatrix(
const TemperatureMatrix &scratch)
: temperature_fe_values(
scratch.temperature_fe_values.get_mapping(),
scratch.temperature_fe_values.get_fe(),
scratch.temperature_fe_values.get_quadrature(),
scratch.temperature_fe_values.get_update_flags())
, phi_T(scratch.phi_T)
, grad_phi_T(scratch.grad_phi_T)
{}
@ update_values
Shape function values.
@ update_JxW_values
Transformed quadrature weights.
@ update_gradients
Shape function gradients.
The final scratch object is used in the assembly of the right hand side of the temperature system. This object is significantly larger than the ones above because a lot more quantities enter the computation of the right hand side of the temperature equation. In particular, the temperature values and gradients of the previous two time steps need to be evaluated at the quadrature points, as well as the velocities and the strain rates (i.e. the symmetric gradients of the velocity) that enter the right hand side as friction heating terms. Despite the number of terms, the following should be rather self explanatory:
template <int dim>
struct TemperatureRHS
{
TemperatureRHS(const TemperatureRHS &data);
std::vector<double> phi_T;
std::vector<Tensor<1, dim>> grad_phi_T;
std::vector<Tensor<1, dim>> old_velocity_values;
std::vector<Tensor<1, dim>> old_old_velocity_values;
std::vector<SymmetricTensor<2, dim>> old_strain_rates;
std::vector<SymmetricTensor<2, dim>> old_old_strain_rates;
std::vector<double> old_temperature_values;
std::vector<double> old_old_temperature_values;
std::vector<Tensor<1, dim>> old_temperature_grads;
std::vector<Tensor<1, dim>> old_old_temperature_grads;
std::vector<double> old_temperature_laplacians;
std::vector<double> old_old_temperature_laplacians;
};
template <int dim>
TemperatureRHS<dim>::TemperatureRHS(
: temperature_fe_values(mapping,
temperature_fe,
quadrature,
, stokes_fe_values(mapping,
stokes_fe,
quadrature,
, phi_T(temperature_fe.n_dofs_per_cell())
, grad_phi_T(temperature_fe.n_dofs_per_cell())
,
old_velocity_values(quadrature.size())
, old_old_velocity_values(quadrature.size())
, old_strain_rates(quadrature.size())
, old_old_strain_rates(quadrature.size())
,
old_temperature_values(quadrature.size())
, old_old_temperature_values(quadrature.size())
, old_temperature_grads(quadrature.size())
, old_old_temperature_grads(quadrature.size())
, old_temperature_laplacians(quadrature.size())
, old_old_temperature_laplacians(quadrature.size())
{}
template <int dim>
TemperatureRHS<dim>::TemperatureRHS(const TemperatureRHS &scratch)
: temperature_fe_values(
scratch.temperature_fe_values.get_mapping(),
scratch.temperature_fe_values.get_fe(),
scratch.temperature_fe_values.get_quadrature(),
scratch.temperature_fe_values.get_update_flags())
, stokes_fe_values(scratch.stokes_fe_values.get_mapping(),
scratch.stokes_fe_values.get_fe(),
scratch.stokes_fe_values.get_quadrature(),
scratch.stokes_fe_values.get_update_flags())
, phi_T(scratch.phi_T)
, grad_phi_T(scratch.grad_phi_T)
,
old_velocity_values(scratch.old_velocity_values)
, old_old_velocity_values(scratch.old_old_velocity_values)
, old_strain_rates(scratch.old_strain_rates)
, old_old_strain_rates(scratch.old_old_strain_rates)
,
old_temperature_values(scratch.old_temperature_values)
, old_old_temperature_values(scratch.old_old_temperature_values)
, old_temperature_grads(scratch.old_temperature_grads)
, old_old_temperature_grads(scratch.old_old_temperature_grads)
, old_temperature_laplacians(scratch.old_temperature_laplacians)
, old_old_temperature_laplacians(scratch.old_old_temperature_laplacians)
{}
}
@ update_hessians
Second derivatives of shape functions.
@ update_quadrature_points
Transformed quadrature points.
The CopyData objects are even simpler than the Scratch objects as all they have to do is to store the results of local computations until they can be copied into the global matrix or vector objects. These structures therefore only need to provide a constructor, a copy operation, and some arrays for local matrix, local vectors and the relation between local and global degrees of freedom (a.k.a. local_dof_indices). Again, we have one such structure for each of the four operations we will parallelize using the WorkStream class:
namespace CopyData
{
template <int dim>
struct StokesPreconditioner
{
StokesPreconditioner(const StokesPreconditioner &data);
StokesPreconditioner &operator=(const StokesPreconditioner &) = default;
std::vector<types::global_dof_index> local_dof_indices;
};
template <int dim>
StokesPreconditioner<dim>::StokesPreconditioner(
: local_matrix(stokes_fe.n_dofs_per_cell(), stokes_fe.n_dofs_per_cell())
, local_dof_indices(stokes_fe.n_dofs_per_cell())
{}
template <int dim>
StokesPreconditioner<dim>::StokesPreconditioner(
const StokesPreconditioner &data)
: local_matrix(data.local_matrix)
, local_dof_indices(data.local_dof_indices)
{}
template <int dim>
struct StokesSystem : public StokesPreconditioner<dim>
{
};
template <int dim>
: StokesPreconditioner<dim>(stokes_fe)
, local_rhs(stokes_fe.n_dofs_per_cell())
{}
template <int dim>
struct TemperatureMatrix
{
std::vector<types::global_dof_index> local_dof_indices;
};
template <int dim>
TemperatureMatrix<dim>::TemperatureMatrix(
: local_mass_matrix(temperature_fe.n_dofs_per_cell(),
temperature_fe.n_dofs_per_cell())
, local_stiffness_matrix(temperature_fe.n_dofs_per_cell(),
temperature_fe.n_dofs_per_cell())
, local_dof_indices(temperature_fe.n_dofs_per_cell())
{}
template <int dim>
struct TemperatureRHS
{
std::vector<types::global_dof_index> local_dof_indices;
};
template <int dim>
TemperatureRHS<dim>::TemperatureRHS(
: local_rhs(temperature_fe.n_dofs_per_cell())
, local_dof_indices(temperature_fe.n_dofs_per_cell())
, matrix_for_bc(temperature_fe.n_dofs_per_cell(),
temperature_fe.n_dofs_per_cell())
{}
}
}
The BoussinesqFlowProblem class template
This is the declaration of the main class. It is very similar to step-31 but there are a number differences we will comment on below.
The top of the class is essentially the same as in step-31, listing the public methods and a set of private functions that do the heavy lifting. Compared to step-31 there are only two additions to this section: the function get_cfl_number() that computes the maximum CFL number over all cells which we then compute the global time step from, and the function get_entropy_variation() that is used in the computation of the entropy stabilization. It is akin to the get_extrapolated_temperature_range() we have used in step-31 for this purpose, but works on the entropy instead of the temperature instead.
template <int dim>
class BoussinesqFlowProblem
{
public:
struct Parameters;
BoussinesqFlowProblem(Parameters ¶meters);
private:
void setup_dofs();
void assemble_stokes_preconditioner();
void build_stokes_preconditioner();
void assemble_stokes_system();
void assemble_temperature_matrix();
void assemble_temperature_system(const double maximal_velocity);
double get_maximal_velocity() const;
double get_cfl_number() const;
double get_entropy_variation(const double average_temperature) const;
std::pair<double, double> get_extrapolated_temperature_range() const;
void solve();
void output_results();
void refine_mesh(const unsigned int max_grid_level);
double compute_viscosity(
const std::vector<double> & old_temperature,
const std::vector<double> & old_old_temperature,
const std::vector<double> & old_temperature_laplacians,
const std::vector<double> & old_old_temperature_laplacians,
const double global_u_infty,
const double global_T_variation,
const double average_temperature,
const double global_entropy_variation,
const double cell_diameter) const;
public:
void run(const Iterator &begin, const typename identity< Iterator >::type &end, Worker worker, Copier copier, const ScratchData &sample_scratch_data, const CopyData &sample_copy_data, const unsigned int queue_length, const unsigned int chunk_size)
The first significant new component is the definition of a struct for the parameters according to the discussion in the introduction. This structure is initialized by reading from a parameter file during construction of this object.
struct Parameters
{
Parameters(const std::string ¶meter_filename);
double end_time;
unsigned int initial_global_refinement;
unsigned int initial_adaptive_refinement;
bool generate_graphical_output;
unsigned int graphical_output_interval;
unsigned int adaptive_refinement_interval;
double stabilization_alpha;
double stabilization_c_R;
double stabilization_beta;
unsigned int stokes_velocity_degree;
bool use_locally_conservative_discretization;
unsigned int temperature_degree;
};
private:
Parameters ¶meters;
The pcout (for parallel std::cout) object is used to simplify writing output: each MPI process can use this to generate output as usual, but since each of these processes will (hopefully) produce the same output it will just be replicated many times over; with the ConditionalOStream class, only the output generated by one MPI process will actually be printed to screen, whereas the output by all the other threads will simply be forgotten.
The following member variables will then again be similar to those in step-31 (and to other tutorial programs). As mentioned in the introduction, we fully distribute computations, so we will have to use the parallel::distributed::Triangulation class (see step-40) but the remainder of these variables is rather standard with two exceptions:
- The
mapping variable is used to denote a higher-order polynomial mapping. As mentioned in the introduction, we use this mapping when forming integrals through quadrature for all cells that are adjacent to either the inner or outer boundaries of our domain where the boundary is curved.
- In a bit of naming confusion, you will notice below that some of the variables from namespace TrilinosWrappers are taken from namespace TrilinosWrappers::MPI (such as the right hand side vectors) whereas others are not (such as the various matrices). This is due to legacy reasons. We will frequently have to query velocities and temperatures at arbitrary quadrature points; consequently, rather than importing ghost information of a vector whenever we need access to degrees of freedom that are relevant locally but owned by another processor, we solve linear systems in parallel but then immediately initialize a vector including ghost entries of the solution for further processing. The various
*_solution vectors are therefore filled immediately after solving their respective linear system in parallel and will always contain values for all locally relevant degrees of freedom; the fully distributed vectors that we obtain from the solution process and that only ever contain the locally owned degrees of freedom are destroyed immediately after the solution process and after we have copied the relevant values into the member variable vectors.
double global_Omega_diameter;
double time_step;
double old_time_step;
unsigned int timestep_number;
std::shared_ptr<TrilinosWrappers::PreconditionAMG> Amg_preconditioner;
std::shared_ptr<TrilinosWrappers::PreconditionJacobi> Mp_preconditioner;
std::shared_ptr<TrilinosWrappers::PreconditionJacobi> T_preconditioner;
bool rebuild_stokes_matrix;
bool rebuild_stokes_preconditioner;
bool rebuild_temperature_matrices;
bool rebuild_temperature_preconditioner;
const ::parallel::distributed::Triangulation< dim, spacedim > * triangulation
The next member variable, computing_timer is used to conveniently account for compute time spent in certain "sections" of the code that are repeatedly entered. For example, we will enter (and leave) sections for Stokes matrix assembly and would like to accumulate the run time spent in this section over all time steps. Every so many time steps as well as at the end of the program (through the destructor of the TimerOutput class) we will then produce a nice summary of the times spent in the different sections into which we categorize the run-time of this program.
After these member variables we have a number of auxiliary functions that have been broken out of the ones listed above. Specifically, there are first three functions that we call from setup_dofs and then the ones that do the assembling of linear systems:
void setup_stokes_matrix(
const std::vector<IndexSet> &stokes_partitioning,
const std::vector<IndexSet> &stokes_relevant_partitioning);
void setup_stokes_preconditioner(
const std::vector<IndexSet> &stokes_partitioning,
const std::vector<IndexSet> &stokes_relevant_partitioning);
void setup_temperature_matrices(
const IndexSet &temperature_partitioning,
const IndexSet &temperature_relevant_partitioning);
Following the task-based parallelization paradigm, we split all the assembly routines into two parts: a first part that can do all the calculations on a certain cell without taking care of other threads, and a second part (which is writing the local data into the global matrices and vectors) which can be entered by only one thread at a time. In order to implement that, we provide functions for each of those two steps for all the four assembly routines that we use in this program. The following eight functions do exactly this:
void local_assemble_stokes_preconditioner(
Assembly::Scratch::StokesPreconditioner<dim> & scratch,
Assembly::CopyData::StokesPreconditioner<dim> & data);
void copy_local_to_global_stokes_preconditioner(
const Assembly::CopyData::StokesPreconditioner<dim> &data);
void local_assemble_stokes_system(
Assembly::Scratch::StokesSystem<dim> & scratch,
Assembly::CopyData::StokesSystem<dim> & data);
void copy_local_to_global_stokes_system(
const Assembly::CopyData::StokesSystem<dim> &data);
void local_assemble_temperature_matrix(
Assembly::Scratch::TemperatureMatrix<dim> & scratch,
Assembly::CopyData::TemperatureMatrix<dim> & data);
void copy_local_to_global_temperature_matrix(
const Assembly::CopyData::TemperatureMatrix<dim> &data);
void local_assemble_temperature_rhs(
const std::pair<double, double> global_T_range,
const double global_max_velocity,
const double global_entropy_variation,
Assembly::Scratch::TemperatureRHS<dim> & scratch,
Assembly::CopyData::TemperatureRHS<dim> & data);
void copy_local_to_global_temperature_rhs(
const Assembly::CopyData::TemperatureRHS<dim> &data);
typename ActiveSelector::active_cell_iterator active_cell_iterator
Finally, we forward declare a member class that we will define later on and that will be used to compute a number of quantities from our solution vectors that we'd like to put into the output files for visualization.
BoussinesqFlowProblem class implementation
BoussinesqFlowProblem::Parameters
Here comes the definition of the parameters for the Stokes problem. We allow to set the end time for the simulation, the level of refinements (both global and adaptive, which in the sum specify what maximum level the cells are allowed to have), and the interval between refinements in the time stepping.
Then, we let the user specify constants for the stabilization parameters (as discussed in the introduction), the polynomial degree for the Stokes velocity space, whether to use the locally conservative discretization based on FE_DGP elements for the pressure or not (FE_Q elements for pressure), and the polynomial degree for the temperature interpolation.
The constructor checks for a valid input file (if not, a file with default parameters for the quantities is written), and eventually parses the parameters.
template <int dim>
BoussinesqFlowProblem<dim>::Parameters::Parameters(
const std::string ¶meter_filename)
: end_time(1e8)
, initial_global_refinement(2)
, initial_adaptive_refinement(2)
, adaptive_refinement_interval(10)
, stabilization_alpha(2)
, stabilization_c_R(0.11)
, stabilization_beta(0.078)
, stokes_velocity_degree(2)
, use_locally_conservative_discretization(true)
, temperature_degree(2)
{
BoussinesqFlowProblem<dim>::Parameters::declare_parameters(prm);
std::ifstream parameter_file(parameter_filename);
if (!parameter_file)
{
parameter_file.close();
std::ofstream parameter_out(parameter_filename);
false,
"Input parameter file <" + parameter_filename +
"> not found. Creating a template file of the same name."));
}
parse_parameters(prm);
}
virtual void parse_input(std::istream &input, const std::string &filename="input file", const std::string &last_line="", const bool skip_undefined=false)
std::ostream & print_parameters(std::ostream &out, const OutputStyle style) const
static ::ExceptionBase & ExcMessage(std::string arg1)
#define AssertThrow(cond, exc)
Next we have a function that declares the parameters that we expect in the input file, together with their data types, default values and a description:
template <int dim>
void BoussinesqFlowProblem<dim>::Parameters::declare_parameters(
{
"1e8",
"The end time of the simulation in years.");
"2",
"The number of global refinement steps performed on "
"the initial coarse mesh, before the problem is first "
"solved there.");
"2",
"The number of adaptive refinement steps performed after "
"initial global refinement.");
"10",
"The number of time steps after which the mesh is to be "
"adapted based on computed error indicators.");
"false",
"Whether graphical output is to be generated or not. "
"You may not want to get graphical output if the number "
"of processors is large.");
"50",
"The number of time steps between each generation of "
"graphical output files.");
{
"2",
"The exponent in the entropy viscosity stabilization.");
"0.11",
"The c_R factor in the entropy viscosity "
"stabilization.");
"0.078",
"The beta factor in the artificial viscosity "
"stabilization. An appropriate value for 2d is 0.052 "
"and 0.078 for 3d.");
}
{
"Stokes velocity polynomial degree",
"2",
"The polynomial degree to use for the velocity variables "
"in the Stokes system.");
"Temperature polynomial degree",
"2",
"The polynomial degree to use for the temperature variable.");
"Use locally conservative discretization",
"true",
"Whether to use a Stokes discretization that is locally "
"conservative at the expense of a larger number of degrees "
"of freedom, or to go with a cheaper discretization "
"that does not locally conserve mass (although it is "
"globally conservative.");
}
}
void declare_entry(const std::string &entry, const std::string &default_value, const Patterns::PatternBase &pattern=Patterns::Anything(), const std::string &documentation="", const bool has_to_be_set=false)
void enter_subsection(const std::string &subsection)
And then we need a function that reads the contents of the ParameterHandler object we get by reading the input file and puts the results into variables that store the values of the parameters we have previously declared:
template <int dim>
void BoussinesqFlowProblem<dim>::Parameters::parse_parameters(
{
initial_global_refinement = prm.
get_integer(
"Initial global refinement");
initial_adaptive_refinement =
adaptive_refinement_interval =
generate_graphical_output = prm.
get_bool(
"Generate graphical output");
graphical_output_interval =
prm.
get_integer(
"Time steps between graphical output");
{
}
{
stokes_velocity_degree =
temperature_degree = prm.
get_integer(
"Temperature polynomial degree");
use_locally_conservative_discretization =
prm.
get_bool(
"Use locally conservative discretization");
}
}
long int get_integer(const std::string &entry_string) const
bool get_bool(const std::string &entry_name) const
double get_double(const std::string &entry_name) const
BoussinesqFlowProblem::BoussinesqFlowProblem
The constructor of the problem is very similar to the constructor in step-31. What is different is the parallel communication: Trilinos uses a message passing interface (MPI) for data distribution. When entering the BoussinesqFlowProblem class, we have to decide how the parallelization is to be done. We choose a rather simple strategy and let all processors that are running the program work together, specified by the communicator MPI_COMM_WORLD. Next, we create the output stream (as we already did in step-18) that only generates output on the first MPI process and is completely forgetful on all others. The implementation of this idea is to check the process number when pcout gets a true argument, and it uses the std::cout stream for output. If we are one processor five, for instance, then we will give a false argument to pcout, which means that the output of that processor will not be printed. With the exception of the mapping object (for which we use polynomials of degree 4) all but the final member variable are exactly the same as in step-31.
This final object, the TimerOutput object, is then told to restrict output to the pcout stream (processor 0), and then we specify that we want to get a summary table at the end of the program which shows us wallclock times (as opposed to CPU times). We will manually also request intermediate summaries every so many time steps in the run() function below.
template <int dim>
BoussinesqFlowProblem<dim>::BoussinesqFlowProblem(Parameters ¶meters_)
: parameters(parameters_)
,
,
global_Omega_diameter(0.)
,
mapping(4)
,
stokes_fe(
FE_Q<dim>(parameters.stokes_velocity_degree),
dim,
(parameters.use_locally_conservative_discretization ?
FE_DGP<dim>(parameters.stokes_velocity_degree - 1)) :
FE_Q<dim>(parameters.stokes_velocity_degree - 1))),
1)
,
,
temperature_fe(parameters.temperature_degree)
,
time_step(0)
, old_time_step(0)
, timestep_number(0)
, rebuild_stokes_matrix(true)
, rebuild_stokes_preconditioner(true)
, rebuild_temperature_matrices(true)
, rebuild_temperature_preconditioner(true)
,
computing_timer(MPI_COMM_WORLD,
pcout,
{}
unsigned int this_mpi_process(const MPI_Comm &mpi_communicator)
The BoussinesqFlowProblem helper functions
BoussinesqFlowProblem::get_maximal_velocity
Except for two small details, the function to compute the global maximum of the velocity is the same as in step-31. The first detail is actually common to all functions that implement loops over all cells in the triangulation: When operating in parallel, each processor can only work on a chunk of cells since each processor only has a certain part of the entire triangulation. This chunk of cells that we want to work on is identified via a so-called subdomain_id, as we also did in step-18. All we need to change is hence to perform the cell-related operations only on cells that are owned by the current process (as opposed to ghost or artificial cells), i.e. for which the subdomain id equals the number of the process ID. Since this is a commonly used operation, there is a shortcut for this operation: we can ask whether the cell is owned by the current processor using cell->is_locally_owned().
The second difference is the way we calculate the maximum value. Before, we could simply have a double variable that we checked against on each quadrature point for each cell. Now, we have to be a bit more careful since each processor only operates on a subset of cells. What we do is to first let each processor calculate the maximum among its cells, and then do a global communication operation Utilities::MPI::max that computes the maximum value among all the maximum values of the individual processors. MPI provides such a call, but it's even simpler to use the respective function in namespace Utilities::MPI using the MPI communicator object since that will do the right thing even if we work without MPI and on a single machine only. The call to Utilities::MPI::max needs two arguments, namely the local maximum (input) and the MPI communicator, which is MPI_COMM_WORLD in this example.
template <int dim>
double BoussinesqFlowProblem<dim>::get_maximal_velocity() const
{
parameters.stokes_velocity_degree);
const unsigned int n_q_points = quadrature_formula.size();
stokes_fe,
quadrature_formula,
std::vector<Tensor<1, dim>> velocity_values(n_q_points);
double max_local_velocity = 0;
for (const auto &cell : stokes_dof_handler.active_cell_iterators())
if (cell->is_locally_owned())
{
fe_values.reinit(cell);
fe_values[velocities].get_function_values(stokes_solution,
velocity_values);
for (unsigned int q = 0; q < n_q_points; ++q)
max_local_velocity =
}
}
double norm(const FEValuesBase< dim > &fe, const ArrayView< const std::vector< Tensor< 1, dim > > > &Du)
T max(const T &t, const MPI_Comm &mpi_communicator)
BoussinesqFlowProblem::get_cfl_number
The next function does something similar, but we now compute the CFL number, i.e., maximal velocity on a cell divided by the cell diameter. This number is necessary to determine the time step size, as we use a semi-explicit time stepping scheme for the temperature equation (see step-31 for a discussion). We compute it in the same way as above: Compute the local maximum over all locally owned cells, then exchange it via MPI to find the global maximum.
template <int dim>
double BoussinesqFlowProblem<dim>::get_cfl_number() const
{
parameters.stokes_velocity_degree);
const unsigned int n_q_points = quadrature_formula.size();
stokes_fe,
quadrature_formula,
std::vector<Tensor<1, dim>> velocity_values(n_q_points);
double max_local_cfl = 0;
for (const auto &cell : stokes_dof_handler.active_cell_iterators())
if (cell->is_locally_owned())
{
fe_values.reinit(cell);
fe_values[velocities].get_function_values(stokes_solution,
velocity_values);
double max_local_velocity = 1
e-10;
for (unsigned int q = 0; q < n_q_points; ++q)
max_local_velocity =
max_local_cfl =
std::max(max_local_cfl, max_local_velocity / cell->diameter());
}
}
BoussinesqFlowProblem::get_entropy_variation
Next comes the computation of the global entropy variation \(\|E(T)-\bar{E}(T)\|_\infty\) where the entropy \(E\) is defined as discussed in the introduction. This is needed for the evaluation of the stabilization in the temperature equation as explained in the introduction. The entropy variation is actually only needed if we use \(\alpha=2\) as a power in the residual computation. The infinity norm is computed by the maxima over quadrature points, as usual in discrete computations.
In order to compute this quantity, we first have to find the space-average \(\bar{E}(T)\) and then evaluate the maximum. However, that means that we would need to perform two loops. We can avoid the overhead by noting that \(\|E(T)-\bar{E}(T)\|_\infty =
\max\big(E_{\textrm{max}}(T)-\bar{E}(T),
\bar{E}(T)-E_{\textrm{min}}(T)\big)\), i.e., the maximum out of the deviation from the average entropy in positive and negative directions. The four quantities we need for the latter formula (maximum entropy, minimum entropy, average entropy, area) can all be evaluated in the same loop over all cells, so we choose this simpler variant.
template <int dim>
double BoussinesqFlowProblem<dim>::get_entropy_variation(
const double average_temperature) const
{
if (parameters.stabilization_alpha != 2)
return 1.;
const QGauss<dim> quadrature_formula(parameters.temperature_degree + 1);
const unsigned int n_q_points = quadrature_formula.size();
quadrature_formula,
std::vector<double> old_temperature_values(n_q_points);
std::vector<double> old_old_temperature_values(n_q_points);
In the two functions above we computed the maximum of numbers that were all non-negative, so we knew that zero was certainly a lower bound. On the other hand, here we need to find the maximum deviation from the average value, i.e., we will need to know the maximal and minimal values of the entropy for which we don't a priori know the sign.
To compute it, we can therefore start with the largest and smallest possible values we can store in a double precision number: The minimum is initialized with a bigger and the maximum with a smaller number than any one that is going to appear. We are then guaranteed that these numbers will be overwritten in the loop on the first cell or, if this processor does not own any cells, in the communication step at the latest. The following loop then computes the minimum and maximum local entropy as well as keeps track of the area/volume of the part of the domain we locally own and the integral over the entropy on it:
entropy_integrated = 0;
for (const auto &cell : temperature_dof_handler.active_cell_iterators())
if (cell->is_locally_owned())
{
fe_values.reinit(cell);
fe_values.get_function_values(old_temperature_solution,
old_temperature_values);
fe_values.get_function_values(old_old_temperature_solution,
old_old_temperature_values);
for (unsigned int q = 0; q < n_q_points; ++q)
{
(old_temperature_values[q] + old_old_temperature_values[q]) / 2;
const double entropy =
((
T - average_temperature) * (
T - average_temperature));
min_entropy =
std::min(min_entropy, entropy);
max_entropy =
std::max(max_entropy, entropy);
area += fe_values.JxW(q);
entropy_integrated += fe_values.JxW(q) * entropy;
}
}
::VectorizedArray< Number, width > min(const ::VectorizedArray< Number, width > &, const ::VectorizedArray< Number, width > &)
Now we only need to exchange data between processors: we need to sum the two integrals (area, entropy_integrated), and get the extrema for maximum and minimum. We could do this through four different data exchanges, but we can it with two: Utilities::MPI::sum also exists in a variant that takes an array of values that are all to be summed up. And we can also utilize the Utilities::MPI::max function by realizing that forming the minimum over the minimal entropies equals forming the negative of the maximum over the negative of the minimal entropies; this maximum can then be combined with forming the maximum over the maximal entropies.
const double local_sums[2] = {entropy_integrated, area},
local_maxima[2] = {-min_entropy, max_entropy};
double global_sums[2], global_maxima[2];
T sum(const T &t, const MPI_Comm &mpi_communicator)
Having computed everything this way, we can then compute the average entropy and find the \(L^\infty\) norm by taking the larger of the deviation of the maximum or minimum from the average:
const double average_entropy = global_sums[0] / global_sums[1];
const double entropy_diff =
std::max(global_maxima[1] - average_entropy,
average_entropy - (-global_maxima[0]));
return entropy_diff;
}
BoussinesqFlowProblem::get_extrapolated_temperature_range
The next function computes the minimal and maximal value of the extrapolated temperature over the entire domain. Again, this is only a slightly modified version of the respective function in step-31. As in the function above, we collect local minima and maxima and then compute the global extrema using the same trick as above.
As already discussed in step-31, the function needs to distinguish between the first and all following time steps because it uses a higher order temperature extrapolation scheme when at least two previous time steps are available.
template <int dim>
std::pair<double, double>
BoussinesqFlowProblem<dim>::get_extrapolated_temperature_range() const
{
parameters.temperature_degree);
const unsigned int n_q_points = quadrature_formula.size();
temperature_fe,
quadrature_formula,
std::vector<double> old_temperature_values(n_q_points);
std::vector<double> old_old_temperature_values(n_q_points);
if (timestep_number != 0)
{
for (const auto &cell : temperature_dof_handler.active_cell_iterators())
if (cell->is_locally_owned())
{
fe_values.reinit(cell);
fe_values.get_function_values(old_temperature_solution,
old_temperature_values);
fe_values.get_function_values(old_old_temperature_solution,
old_old_temperature_values);
for (unsigned int q = 0; q < n_q_points; ++q)
{
const double temperature =
(1. + time_step / old_time_step) *
old_temperature_values[q] -
time_step / old_time_step * old_old_temperature_values[q];
min_local_temperature =
std::min(min_local_temperature, temperature);
max_local_temperature =
std::max(max_local_temperature, temperature);
}
}
}
else
{
for (const auto &cell : temperature_dof_handler.active_cell_iterators())
if (cell->is_locally_owned())
{
fe_values.reinit(cell);
fe_values.get_function_values(old_temperature_solution,
old_temperature_values);
for (unsigned int q = 0; q < n_q_points; ++q)
{
const double temperature = old_temperature_values[q];
min_local_temperature =
std::min(min_local_temperature, temperature);
max_local_temperature =
std::max(max_local_temperature, temperature);
}
}
}
double local_extrema[2] = {-min_local_temperature, max_local_temperature};
double global_extrema[2];
return std::make_pair(-global_extrema[0], global_extrema[1]);
}
BoussinesqFlowProblem::compute_viscosity
The function that calculates the viscosity is purely local and so needs no communication at all. It is mostly the same as in step-31 but with an updated formulation of the viscosity if \(\alpha=2\) is chosen:
template <int dim>
double BoussinesqFlowProblem<dim>::compute_viscosity(
const std::vector<double> & old_temperature,
const std::vector<double> & old_old_temperature,
const std::vector<double> & old_temperature_laplacians,
const std::vector<double> & old_old_temperature_laplacians,
const double global_u_infty,
const double global_T_variation,
const double average_temperature,
const double global_entropy_variation,
const double cell_diameter) const
{
if (global_u_infty == 0)
return 5
e-3 * cell_diameter;
const unsigned int n_q_points = old_temperature.size();
double max_residual = 0;
double max_velocity = 0;
for (unsigned int q = 0; q < n_q_points; ++q)
{
(old_velocity_values[q] + old_old_velocity_values[q]) / 2;
(old_strain_rates[q] + old_old_strain_rates[q]) / 2;
const double T = (old_temperature[q] + old_old_temperature[q]) / 2;
const double dT_dt =
(old_temperature[q] - old_old_temperature[q]) / old_time_step;
const double u_grad_T =
u * (old_temperature_grads[q] + old_old_temperature_grads[q]) / 2;
const double kappa_Delta_T =
EquationData::kappa *
(old_temperature_laplacians[q] + old_old_temperature_laplacians[q]) /
2;
((EquationData::radiogenic_heating * EquationData::density(
T) +
2 * EquationData::eta * strain_rate * strain_rate) /
(EquationData::density(
T) * EquationData::specific_heat));
double residual =
std::abs(dT_dt + u_grad_T - kappa_Delta_T -
gamma);
if (parameters.stabilization_alpha == 2)
residual *=
std::abs(
T - average_temperature);
max_residual =
std::max(residual, max_residual);
}
const double max_viscosity =
(parameters.stabilization_beta * max_velocity * cell_diameter);
if (timestep_number == 0)
return max_viscosity;
else
{
double entropy_viscosity;
if (parameters.stabilization_alpha == 2)
entropy_viscosity =
(parameters.stabilization_c_R * cell_diameter * cell_diameter *
max_residual / global_entropy_variation);
else
entropy_viscosity =
(parameters.stabilization_c_R * cell_diameter *
global_Omega_diameter * max_velocity * max_residual /
(global_u_infty * global_T_variation));
return std::min(max_viscosity, entropy_viscosity);
}
}
#define Assert(cond, exc)
static ::ExceptionBase & ExcInternalError()
long double gamma(const unsigned int n)
::VectorizedArray< Number, width > sqrt(const ::VectorizedArray< Number, width > &)
::VectorizedArray< Number, width > abs(const ::VectorizedArray< Number, width > &)
The BoussinesqFlowProblem setup functions
The following three functions set up the Stokes matrix, the matrix used for the Stokes preconditioner, and the temperature matrix. The code is mostly the same as in step-31, but it has been broken out into three functions of their own for simplicity.
The main functional difference between the code here and that in step-31 is that the matrices we want to set up are distributed across multiple processors. Since we still want to build up the sparsity pattern first for efficiency reasons, we could continue to build the entire sparsity pattern as a BlockDynamicSparsityPattern, as we did in step-31. However, that would be inefficient: every processor would build the same sparsity pattern, but only initialize a small part of the matrix using it. It also violates the principle that every processor should only work on those cells it owns (and, if necessary the layer of ghost cells around it).
Rather, we use an object of type TrilinosWrappers::BlockSparsityPattern, which is (obviously) a wrapper around a sparsity pattern object provided by Trilinos. The advantage is that the Trilinos sparsity pattern class can communicate across multiple processors: if this processor fills in all the nonzero entries that result from the cells it owns, and every other processor does so as well, then at the end after some MPI communication initiated by the compress() call, we will have the globally assembled sparsity pattern available with which the global matrix can be initialized.
There is one important aspect when initializing Trilinos sparsity patterns in parallel: In addition to specifying the locally owned rows and columns of the matrices via the stokes_partitioning index set, we also supply information about all the rows we are possibly going to write into when assembling on a certain processor. The set of locally relevant rows contains all such rows (possibly also a few unnecessary ones, but it is difficult to find the exact row indices before actually getting indices on all cells and resolving constraints). This additional information allows to exactly determine the structure for the off-processor data found during assembly. While Trilinos matrices are able to collect this information on the fly as well (when initializing them from some other reinit method), it is less efficient and leads to problems when assembling matrices with multiple threads. In this program, we pessimistically assume that only one processor at a time can write into the matrix while assembly (whereas the computation is parallel), which is fine for Trilinos matrices. In practice, one can do better by hinting WorkStream at cells that do not share vertices, allowing for parallelism among those cells (see the graph coloring algorithms and WorkStream with colored iterators argument). However, that only works when only one MPI processor is present because Trilinos' internal data structures for accumulating off-processor data on the fly are not thread safe. With the initialization presented here, there is no such problem and one could safely introduce graph coloring for this algorithm.
The only other change we need to make is to tell the DoFTools::make_sparsity_pattern() function that it is only supposed to work on a subset of cells, namely the ones whose subdomain_id equals the number of the current processor, and to ignore all other cells.
This strategy is replicated across all three of the following functions.
Note that Trilinos matrices store the information contained in the sparsity patterns, so we can safely release the sp variable once the matrix has been given the sparsity structure.
template <int dim>
void BoussinesqFlowProblem<dim>::setup_stokes_matrix(
const std::vector<IndexSet> &stokes_partitioning,
const std::vector<IndexSet> &stokes_relevant_partitioning)
{
stokes_matrix.clear();
stokes_partitioning,
stokes_relevant_partitioning,
MPI_COMM_WORLD);
for (unsigned int c = 0; c < dim + 1; ++c)
for (
unsigned int d = 0;
d < dim + 1; ++
d)
if (!((c == dim) && (
d == dim)))
else
coupling,
sp,
stokes_constraints,
false,
MPI_COMM_WORLD));
sp.compress();
stokes_matrix.reinit(sp);
}
template <int dim>
void BoussinesqFlowProblem<dim>::setup_stokes_preconditioner(
const std::vector<IndexSet> &stokes_partitioning,
const std::vector<IndexSet> &stokes_relevant_partitioning)
{
Amg_preconditioner.reset();
Mp_preconditioner.reset();
stokes_preconditioner_matrix.clear();
stokes_partitioning,
stokes_relevant_partitioning,
MPI_COMM_WORLD);
for (unsigned int c = 0; c < dim + 1; ++c)
for (
unsigned int d = 0;
d < dim + 1; ++
d)
else
coupling,
sp,
stokes_constraints,
false,
MPI_COMM_WORLD));
sp.compress();
stokes_preconditioner_matrix.reinit(sp);
}
template <int dim>
void BoussinesqFlowProblem<dim>::setup_temperature_matrices(
const IndexSet &temperature_partitioner,
const IndexSet &temperature_relevant_partitioner)
{
T_preconditioner.reset();
temperature_mass_matrix.clear();
temperature_stiffness_matrix.clear();
temperature_matrix.clear();
temperature_partitioner,
temperature_relevant_partitioner,
MPI_COMM_WORLD);
sp,
temperature_constraints,
false,
MPI_COMM_WORLD));
sp.compress();
temperature_matrix.reinit(sp);
temperature_mass_matrix.reinit(sp);
temperature_stiffness_matrix.reinit(sp);
}
void make_sparsity_pattern(const DoFHandler< dim, spacedim > &dof_handler, SparsityPatternType &sparsity_pattern, const AffineConstraints< number > &constraints=AffineConstraints< number >(), const bool keep_constrained_dofs=true, const types::subdomain_id subdomain_id=numbers::invalid_subdomain_id)
SymmetricTensor< 2, dim, Number > d(const Tensor< 2, dim, Number > &F, const Tensor< 2, dim, Number > &dF_dt)
The remainder of the setup function (after splitting out the three functions above) mostly has to deal with the things we need to do for parallelization across processors. Because setting all of this up is a significant compute time expense of the program, we put everything we do here into a timer group so that we can get summary information about the fraction of time spent in this part of the program at its end.
At the top as usual we enumerate degrees of freedom and sort them by component/block, followed by writing their numbers to the screen from processor zero. The DoFHandler::distributed_dofs() function, when applied to a parallel::distributed::Triangulation object, sorts degrees of freedom in such a way that all degrees of freedom associated with subdomain zero come before all those associated with subdomain one, etc. For the Stokes part, this entails, however, that velocities and pressures become intermixed, but this is trivially solved by sorting again by blocks; it is worth noting that this latter operation leaves the relative ordering of all velocities and pressures alone, i.e. within the velocity block we will still have all those associated with subdomain zero before all velocities associated with subdomain one, etc. This is important since we store each of the blocks of this matrix distributed across all processors and want this to be done in such a way that each processor stores that part of the matrix that is roughly equal to the degrees of freedom located on those cells that it will actually work on.
When printing the numbers of degrees of freedom, note that these numbers are going to be large if we use many processors. Consequently, we let the stream put a comma separator in between every three digits. The state of the stream, using the locale, is saved from before to after this operation. While slightly opaque, the code works because the default locale (which we get using the constructor call std::locale("")) implies printing numbers with a comma separator for every third digit (i.e., thousands, millions, billions).
In this function as well as many below, we measure how much time we spend here and collect that in a section called "Setup dof
systems" across function invocations. This is done using an TimerOutput::Scope object that gets a timer going in the section with above name of the computing_timer object upon construction of the local variable; the timer is stopped again when the destructor of the timing_section variable is called. This, of course, happens either at the end of the function, or if we leave the function through a return statement or when an exception is thrown somewhere – in other words, whenever we leave this function in any way. The use of such "scope" objects therefore makes sure that we do not have to manually add code that tells the timer to stop at every location where this function may be left.
template <int dim>
void BoussinesqFlowProblem<dim>::setup_dofs()
{
stokes_dof_handler.distribute_dofs(stokes_fe);
std::vector<unsigned int> stokes_sub_blocks(dim + 1, 0);
stokes_sub_blocks[dim] = 1;
temperature_dof_handler.distribute_dofs(temperature_fe);
const std::vector<types::global_dof_index> stokes_dofs_per_block =
const unsigned int n_u = stokes_dofs_per_block[0],
n_p = stokes_dofs_per_block[1],
n_T = temperature_dof_handler.n_dofs();
std::locale s = pcout.get_stream().getloc();
pcout.get_stream().imbue(std::locale(""));
pcout <<
"Number of active cells: " <<
triangulation.n_global_active_cells()
<<
" (on " <<
triangulation.n_levels() <<
" levels)" << std::endl
<< "Number of degrees of freedom: " << n_u + n_p + n_T << " (" << n_u
<< '+' << n_p << '+' << n_T << ')' << std::endl
<< std::endl;
pcout.get_stream().imbue(s);
void component_wise(DoFHandler< dim, spacedim > &dof_handler, const std::vector< unsigned int > &target_component=std::vector< unsigned int >())
After this, we have to set up the various partitioners (of type IndexSet, see the introduction) that describe which parts of each matrix or vector will be stored where, then call the functions that actually set up the matrices, and at the end also resize the various vectors we keep around in this program.
std::vector<IndexSet> stokes_partitioning, stokes_relevant_partitioning;
temperature_relevant_partitioning(n_T);
{
IndexSet stokes_index_set = stokes_dof_handler.locally_owned_dofs();
stokes_partitioning.push_back(stokes_index_set.
get_view(0, n_u));
stokes_partitioning.push_back(stokes_index_set.
get_view(n_u, n_u + n_p));
stokes_relevant_set);
stokes_relevant_partitioning.push_back(
stokes_relevant_partitioning.push_back(
stokes_relevant_set.
get_view(n_u, n_u + n_p));
temperature_partitioning = temperature_dof_handler.locally_owned_dofs();
temperature_dof_handler, temperature_relevant_partitioning);
}
IndexSet get_view(const size_type begin, const size_type end) const
Following this, we can compute constraints for the solution vectors, including hanging node constraints and homogeneous and inhomogeneous boundary values for the Stokes and temperature fields. Note that as for everything else, the constraint objects can not hold all constraints on every processor. Rather, each processor needs to store only those that are actually necessary for correctness given that it only assembles linear systems on cells it owns. As discussed in the this paper, the set of constraints we need to know about is exactly the set of constraints on all locally relevant degrees of freedom, so this is what we use to initialize the constraint objects.
{
stokes_constraints.clear();
stokes_constraints.reinit(stokes_relevant_set);
stokes_constraints);
stokes_dof_handler,
0,
stokes_constraints,
stokes_fe.component_mask(velocity_components));
std::set<types::boundary_id> no_normal_flux_boundaries;
no_normal_flux_boundaries.insert(1);
0,
no_normal_flux_boundaries,
stokes_constraints,
mapping);
stokes_constraints.close();
}
{
temperature_constraints.clear();
temperature_constraints.reinit(temperature_relevant_partitioning);
temperature_constraints);
temperature_dof_handler,
0,
EquationData::TemperatureInitialValues<dim>(),
temperature_constraints);
temperature_dof_handler,
1,
EquationData::TemperatureInitialValues<dim>(),
temperature_constraints);
temperature_constraints.close();
}
void compute_no_normal_flux_constraints(const DoFHandler< dim, spacedim > &dof_handler, const unsigned int first_vector_component, const std::set< types::boundary_id > &boundary_ids, AffineConstraints< double > &constraints, const Mapping< dim, spacedim > &mapping=(ReferenceCells::get_hypercube< dim >() .template get_default_linear_mapping< dim, spacedim >()))
void make_hanging_node_constraints(const DoFHandler< dim, spacedim > &dof_handler, AffineConstraints< number > &constraints)
All this done, we can then initialize the various matrix and vector objects to their proper sizes. At the end, we also record that all matrices and preconditioners have to be re-computed at the beginning of the next time step. Note how we initialize the vectors for the Stokes and temperature right hand sides: These are writable vectors (last boolean argument set to true) that have the correct one-to-one partitioning of locally owned elements but are still given the relevant partitioning for means of figuring out the vector entries that are going to be set right away. As for matrices, this allows for writing local contributions into the vector with multiple threads (always assuming that the same vector entry is not accessed by multiple threads at the same time). The other vectors only allow for read access of individual elements, including ghosts, but are not suitable for solvers.
setup_stokes_matrix(stokes_partitioning, stokes_relevant_partitioning);
setup_stokes_preconditioner(stokes_partitioning,
stokes_relevant_partitioning);
setup_temperature_matrices(temperature_partitioning,
temperature_relevant_partitioning);
stokes_rhs.reinit(stokes_partitioning,
stokes_relevant_partitioning,
MPI_COMM_WORLD,
true);
stokes_solution.reinit(stokes_relevant_partitioning, MPI_COMM_WORLD);
old_stokes_solution.reinit(stokes_solution);
temperature_rhs.reinit(temperature_partitioning,
temperature_relevant_partitioning,
MPI_COMM_WORLD,
true);
temperature_solution.reinit(temperature_relevant_partitioning,
MPI_COMM_WORLD);
old_temperature_solution.reinit(temperature_solution);
old_old_temperature_solution.reinit(temperature_solution);
rebuild_stokes_matrix = true;
rebuild_stokes_preconditioner = true;
rebuild_temperature_matrices = true;
rebuild_temperature_preconditioner = true;
}
The BoussinesqFlowProblem assembly functions
Following the discussion in the introduction and in the Parallel computing with multiple processors accessing shared memory module, we split the assembly functions into different parts:
-
The local calculations of matrices and right hand sides, given a certain cell as input (these functions are named local_assemble_* below). The resulting function is, in other words, essentially the body of the loop over all cells in step-31. Note, however, that these functions store the result from the local calculations in variables of classes from the CopyData namespace.
-
These objects are then given to the second step which writes the local data into the global data structures (these functions are named copy_local_to_global_* below). These functions are pretty trivial.
-
These two subfunctions are then used in the respective assembly routine (called
assemble_* below), where a WorkStream object is set up and runs over all the cells that belong to the processor's subdomain.
Stokes preconditioner assembly
Let us start with the functions that builds the Stokes preconditioner. The first two of these are pretty trivial, given the discussion above. Note in particular that the main point in using the scratch data object is that we want to avoid allocating any objects on the free space each time we visit a new cell. As a consequence, the assembly function below only has automatic local variables, and everything else is accessed through the scratch data object, which is allocated only once before we start the loop over all cells:
template <int dim>
void BoussinesqFlowProblem<dim>::local_assemble_stokes_preconditioner(
Assembly::Scratch::StokesPreconditioner<dim> & scratch,
Assembly::CopyData::StokesPreconditioner<dim> & data)
{
const unsigned int dofs_per_cell = stokes_fe.n_dofs_per_cell();
const unsigned int n_q_points =
scratch.stokes_fe_values.n_quadrature_points;
scratch.stokes_fe_values.reinit(cell);
cell->get_dof_indices(data.local_dof_indices);
data.local_matrix = 0;
for (unsigned int q = 0; q < n_q_points; ++q)
{
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
scratch.grad_phi_u[k] =
scratch.stokes_fe_values[velocities].gradient(k, q);
scratch.phi_p[k] = scratch.stokes_fe_values[pressure].value(k, q);
}
for (unsigned int i = 0; i < dofs_per_cell; ++i)
for (unsigned int j = 0; j < dofs_per_cell; ++j)
data.local_matrix(i, j) +=
(EquationData::eta *
(1. / EquationData::eta) * EquationData::pressure_scaling *
EquationData::pressure_scaling *
(scratch.phi_p[i] * scratch.phi_p[j])) *
scratch.stokes_fe_values.JxW(q);
}
}
template <int dim>
void BoussinesqFlowProblem<dim>::copy_local_to_global_stokes_preconditioner(
const Assembly::CopyData::StokesPreconditioner<dim> &data)
{
stokes_constraints.distribute_local_to_global(data.local_matrix,
data.local_dof_indices,
stokes_preconditioner_matrix);
}
constexpr ProductType< Number, OtherNumber >::type scalar_product(const SymmetricTensor< 2, dim, Number > &t1, const SymmetricTensor< 2, dim, OtherNumber > &t2)
Now for the function that actually puts things together, using the WorkStream functions. WorkStream::run needs a start and end iterator to enumerate the cells it is supposed to work on. Typically, one would use DoFHandler::begin_active() and DoFHandler::end() for that but here we actually only want the subset of cells that in fact are owned by the current processor. This is where the FilteredIterator class comes into play: you give it a range of cells and it provides an iterator that only iterates over that subset of cells that satisfy a certain predicate (a predicate is a function of one argument that either returns true or false). The predicate we use here is IteratorFilters::LocallyOwnedCell, i.e., it returns true exactly if the cell is owned by the current processor. The resulting iterator range is then exactly what we need.
With this obstacle out of the way, we call the WorkStream::run function with this set of cells, scratch and copy objects, and with pointers to two functions: the local assembly and copy-local-to-global function. These functions need to have very specific signatures: three arguments in the first and one argument in the latter case (see the documentation of the WorkStream::run function for the meaning of these arguments). Note how we use a lambda functions to create a function object that satisfies this requirement. It uses function arguments for the local assembly function that specify cell, scratch data, and copy data, as well as function argument for the copy function that expects the data to be written into the global matrix (also see the discussion in step-13's assemble_linear_system() function). On the other hand, the implicit zeroth argument of member functions (namely the this pointer of the object on which that member function is to operate on) is bound to the this pointer of the current function and is captured. The WorkStream::run function, as a consequence, does not need to know anything about the object these functions work on.
When the WorkStream is executed, it will create several local assembly routines of the first kind for several cells and let some available processors work on them. The function that needs to be synchronized, i.e., the write operation into the global matrix, however, is executed by only one thread at a time in the prescribed order. Of course, this only holds for the parallelization on a single MPI process. Different MPI processes will have their own WorkStream objects and do that work completely independently (and in different memory spaces). In a distributed calculation, some data will accumulate at degrees of freedom that are not owned by the respective processor. It would be inefficient to send data around every time we encounter such a dof. What happens instead is that the Trilinos sparse matrix will keep that data and send it to the owner at the end of assembly, by calling the compress() command.
template <int dim>
void BoussinesqFlowProblem<dim>::assemble_stokes_preconditioner()
{
stokes_preconditioner_matrix = 0;
const QGauss<dim> quadrature_formula(parameters.stokes_velocity_degree + 1);
using CellFilter =
auto worker =
Assembly::Scratch::StokesPreconditioner<dim> & scratch,
Assembly::CopyData::StokesPreconditioner<dim> & data) {
this->local_assemble_stokes_preconditioner(cell, scratch, data);
};
auto copier =
[this](const Assembly::CopyData::StokesPreconditioner<dim> &data) {
this->copy_local_to_global_stokes_preconditioner(data);
};
stokes_dof_handler.begin_active()),
stokes_dof_handler.end()),
worker,
copier,
Assembly::Scratch::StokesPreconditioner<dim>(
stokes_fe,
quadrature_formula,
mapping,
Assembly::CopyData::StokesPreconditioner<dim>(stokes_fe));
}
void run(const std::vector< std::vector< Iterator > > &colored_iterators, Worker worker, Copier copier, const ScratchData &sample_scratch_data, const CopyData &sample_copy_data, const unsigned int queue_length=2 *MultithreadInfo::n_threads(), const unsigned int chunk_size=8)
The final function in this block initiates assembly of the Stokes preconditioner matrix and then in fact builds the Stokes preconditioner. It is mostly the same as in the serial case. The only difference to step-31 is that we use a Jacobi preconditioner for the pressure mass matrix instead of IC, as discussed in the introduction.
template <int dim>
void BoussinesqFlowProblem<dim>::build_stokes_preconditioner()
{
if (rebuild_stokes_preconditioner == false)
return;
" Build Stokes preconditioner");
pcout << " Rebuilding Stokes preconditioner..." << std::flush;
assemble_stokes_preconditioner();
std::vector<std::vector<bool>> constant_modes;
stokes_fe.component_mask(
velocity_components),
constant_modes);
Mp_preconditioner =
std::make_shared<TrilinosWrappers::PreconditionJacobi>();
Amg_preconditioner = std::make_shared<TrilinosWrappers::PreconditionAMG>();
Mp_preconditioner->initialize(stokes_preconditioner_matrix.block(1, 1));
Amg_preconditioner->initialize(stokes_preconditioner_matrix.block(0, 0),
Amg_data);
rebuild_stokes_preconditioner = false;
pcout << std::endl;
}
std::vector< std::vector< bool > > constant_modes
double aggregation_threshold
unsigned int smoother_sweeps
bool higher_order_elements
Stokes system assembly
The next three functions implement the assembly of the Stokes system, again split up into a part performing local calculations, one for writing the local data into the global matrix and vector, and one for actually running the loop over all cells with the help of the WorkStream class. Note that the assembly of the Stokes matrix needs only to be done in case we have changed the mesh. Otherwise, just the (temperature-dependent) right hand side needs to be calculated here. Since we are working with distributed matrices and vectors, we have to call the respective compress() functions in the end of the assembly in order to send non-local data to the owner process.
template <int dim>
void BoussinesqFlowProblem<dim>::local_assemble_stokes_system(
Assembly::Scratch::StokesSystem<dim> & scratch,
Assembly::CopyData::StokesSystem<dim> & data)
{
const unsigned int dofs_per_cell =
scratch.stokes_fe_values.get_fe().n_dofs_per_cell();
const unsigned int n_q_points =
scratch.stokes_fe_values.n_quadrature_points;
scratch.stokes_fe_values.reinit(cell);
&
triangulation, cell->level(), cell->index(), &temperature_dof_handler);
scratch.temperature_fe_values.reinit(temperature_cell);
if (rebuild_stokes_matrix)
data.local_matrix = 0;
data.local_rhs = 0;
scratch.temperature_fe_values.get_function_values(
old_temperature_solution, scratch.old_temperature_values);
for (unsigned int q = 0; q < n_q_points; ++q)
{
const double old_temperature = scratch.old_temperature_values[q];
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
scratch.phi_u[k] = scratch.stokes_fe_values[velocities].value(k, q);
if (rebuild_stokes_matrix)
{
scratch.grads_phi_u[k] =
scratch.stokes_fe_values[velocities].symmetric_gradient(k, q);
scratch.div_phi_u[k] =
scratch.stokes_fe_values[velocities].divergence(k, q);
scratch.phi_p[k] =
scratch.stokes_fe_values[pressure].value(k, q);
}
}
if (rebuild_stokes_matrix == true)
for (unsigned int i = 0; i < dofs_per_cell; ++i)
for (unsigned int j = 0; j < dofs_per_cell; ++j)
data.local_matrix(i, j) +=
(EquationData::eta * 2 *
(scratch.grads_phi_u[i] * scratch.grads_phi_u[j]) -
(EquationData::pressure_scaling * scratch.div_phi_u[i] *
scratch.phi_p[j]) -
(EquationData::pressure_scaling * scratch.phi_p[i] *
scratch.div_phi_u[j])) *
scratch.stokes_fe_values.JxW(q);
scratch.stokes_fe_values.quadrature_point(q));
for (unsigned int i = 0; i < dofs_per_cell; ++i)
data.local_rhs(i) += (EquationData::density(old_temperature) *
gravity * scratch.phi_u[i]) *
scratch.stokes_fe_values.JxW(q);
}
cell->get_dof_indices(data.local_dof_indices);
}
template <int dim>
void BoussinesqFlowProblem<dim>::copy_local_to_global_stokes_system(
const Assembly::CopyData::StokesSystem<dim> &data)
{
if (rebuild_stokes_matrix == true)
stokes_constraints.distribute_local_to_global(data.local_matrix,
data.local_rhs,
data.local_dof_indices,
stokes_matrix,
stokes_rhs);
else
stokes_constraints.distribute_local_to_global(data.local_rhs,
data.local_dof_indices,
stokes_rhs);
}
template <int dim>
void BoussinesqFlowProblem<dim>::assemble_stokes_system()
{
" Assemble Stokes system");
if (rebuild_stokes_matrix == true)
stokes_matrix = 0;
stokes_rhs = 0;
const QGauss<dim> quadrature_formula(parameters.stokes_velocity_degree + 1);
using CellFilter =
stokes_dof_handler.begin_active()),
Assembly::Scratch::StokesSystem<dim> & scratch,
Assembly::CopyData::StokesSystem<dim> & data) {
this->local_assemble_stokes_system(cell, scratch, data);
},
[this](const Assembly::CopyData::StokesSystem<dim> &data) {
this->copy_local_to_global_stokes_system(data);
},
Assembly::Scratch::StokesSystem<dim>(
stokes_fe,
mapping,
quadrature_formula,
temperature_fe,
Assembly::CopyData::StokesSystem<dim>(stokes_fe));
if (rebuild_stokes_matrix == true)
rebuild_stokes_matrix = false;
pcout << std::endl;
}
Temperature matrix assembly
The task to be performed by the next three functions is to calculate a mass matrix and a Laplace matrix on the temperature system. These will be combined in order to yield the semi-implicit time stepping matrix that consists of the mass matrix plus a time step-dependent weight factor times the Laplace matrix. This function is again essentially the body of the loop over all cells from step-31.
The two following functions perform similar services as the ones above.
template <int dim>
void BoussinesqFlowProblem<dim>::local_assemble_temperature_matrix(
Assembly::Scratch::TemperatureMatrix<dim> & scratch,
Assembly::CopyData::TemperatureMatrix<dim> & data)
{
const unsigned int dofs_per_cell =
scratch.temperature_fe_values.get_fe().n_dofs_per_cell();
const unsigned int n_q_points =
scratch.temperature_fe_values.n_quadrature_points;
scratch.temperature_fe_values.reinit(cell);
cell->get_dof_indices(data.local_dof_indices);
data.local_mass_matrix = 0;
data.local_stiffness_matrix = 0;
for (unsigned int q = 0; q < n_q_points; ++q)
{
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
scratch.grad_phi_T[k] =
scratch.temperature_fe_values.shape_grad(k, q);
scratch.phi_T[k] = scratch.temperature_fe_values.shape_value(k, q);
}
for (unsigned int i = 0; i < dofs_per_cell; ++i)
for (unsigned int j = 0; j < dofs_per_cell; ++j)
{
data.local_mass_matrix(i, j) +=
(scratch.phi_T[i] * scratch.phi_T[j] *
scratch.temperature_fe_values.JxW(q));
data.local_stiffness_matrix(i, j) +=
(EquationData::kappa * scratch.grad_phi_T[i] *
scratch.grad_phi_T[j] * scratch.temperature_fe_values.JxW(q));
}
}
}
template <int dim>
void BoussinesqFlowProblem<dim>::copy_local_to_global_temperature_matrix(
const Assembly::CopyData::TemperatureMatrix<dim> &data)
{
temperature_constraints.distribute_local_to_global(data.local_mass_matrix,
data.local_dof_indices,
temperature_mass_matrix);
temperature_constraints.distribute_local_to_global(
data.local_stiffness_matrix,
data.local_dof_indices,
temperature_stiffness_matrix);
}
template <int dim>
void BoussinesqFlowProblem<dim>::assemble_temperature_matrix()
{
if (rebuild_temperature_matrices == false)
return;
" Assemble temperature matrices");
temperature_mass_matrix = 0;
temperature_stiffness_matrix = 0;
const QGauss<dim> quadrature_formula(parameters.temperature_degree + 2);
using CellFilter =
temperature_dof_handler.begin_active()),
temperature_dof_handler.end()),
Assembly::Scratch::TemperatureMatrix<dim> & scratch,
Assembly::CopyData::TemperatureMatrix<dim> & data) {
this->local_assemble_temperature_matrix(cell, scratch, data);
},
[this](const Assembly::CopyData::TemperatureMatrix<dim> &data) {
this->copy_local_to_global_temperature_matrix(data);
},
Assembly::Scratch::TemperatureMatrix<dim>(temperature_fe,
mapping,
quadrature_formula),
Assembly::CopyData::TemperatureMatrix<dim>(temperature_fe));
rebuild_temperature_matrices = false;
rebuild_temperature_preconditioner = true;
}
Temperature right hand side assembly
This is the last assembly function. It calculates the right hand side of the temperature system, which includes the convection and the stabilization terms. It includes a lot of evaluations of old solutions at the quadrature points (which are necessary for calculating the artificial viscosity of stabilization), but is otherwise similar to the other assembly functions. Notice, once again, how we resolve the dilemma of having inhomogeneous boundary conditions, by just making a right hand side at this point (compare the comments for the project() function above): We create some matrix columns with exactly the values that would be entered for the temperature stiffness matrix, in case we have inhomogeneously constrained dofs. That will account for the correct balance of the right hand side vector with the matrix system of temperature.
template <int dim>
void BoussinesqFlowProblem<dim>::local_assemble_temperature_rhs(
const std::pair<double, double> global_T_range,
const double global_max_velocity,
const double global_entropy_variation,
Assembly::Scratch::TemperatureRHS<dim> & scratch,
Assembly::CopyData::TemperatureRHS<dim> & data)
{
const bool use_bdf2_scheme = (timestep_number != 0);
const unsigned int dofs_per_cell =
scratch.temperature_fe_values.get_fe().n_dofs_per_cell();
const unsigned int n_q_points =
scratch.temperature_fe_values.n_quadrature_points;
data.local_rhs = 0;
data.matrix_for_bc = 0;
cell->get_dof_indices(data.local_dof_indices);
scratch.temperature_fe_values.
reinit(cell);
&
triangulation, cell->level(), cell->index(), &stokes_dof_handler);
scratch.stokes_fe_values.reinit(stokes_cell);
scratch.temperature_fe_values.get_function_values(
old_temperature_solution, scratch.old_temperature_values);
scratch.temperature_fe_values.get_function_values(
old_old_temperature_solution, scratch.old_old_temperature_values);
scratch.temperature_fe_values.get_function_gradients(
old_temperature_solution, scratch.old_temperature_grads);
scratch.temperature_fe_values.get_function_gradients(
old_old_temperature_solution, scratch.old_old_temperature_grads);
scratch.temperature_fe_values.get_function_laplacians(
old_temperature_solution, scratch.old_temperature_laplacians);
scratch.temperature_fe_values.get_function_laplacians(
old_old_temperature_solution, scratch.old_old_temperature_laplacians);
scratch.stokes_fe_values[velocities].get_function_values(
stokes_solution, scratch.old_velocity_values);
scratch.stokes_fe_values[velocities].get_function_values(
old_stokes_solution, scratch.old_old_velocity_values);
scratch.stokes_fe_values[velocities].get_function_symmetric_gradients(
stokes_solution, scratch.old_strain_rates);
scratch.stokes_fe_values[velocities].get_function_symmetric_gradients(
old_stokes_solution, scratch.old_old_strain_rates);
const double nu =
compute_viscosity(scratch.old_temperature_values,
scratch.old_old_temperature_values,
scratch.old_temperature_grads,
scratch.old_old_temperature_grads,
scratch.old_temperature_laplacians,
scratch.old_old_temperature_laplacians,
scratch.old_velocity_values,
scratch.old_old_velocity_values,
scratch.old_strain_rates,
scratch.old_old_strain_rates,
global_max_velocity,
global_T_range.second - global_T_range.first,
0.5 * (global_T_range.second + global_T_range.first),
global_entropy_variation,
cell->diameter());
for (unsigned int q = 0; q < n_q_points; ++q)
{
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
scratch.phi_T[k] = scratch.temperature_fe_values.shape_value(k, q);
scratch.grad_phi_T[k] =
scratch.temperature_fe_values.shape_grad(k, q);
}
const double T_term_for_rhs =
(use_bdf2_scheme ?
(scratch.old_temperature_values[q] *
(1 + time_step / old_time_step) -
scratch.old_old_temperature_values[q] * (time_step * time_step) /
(old_time_step * (time_step + old_time_step))) :
scratch.old_temperature_values[q]);
const double ext_T =
(use_bdf2_scheme ? (scratch.old_temperature_values[q] *
(1 + time_step / old_time_step) -
scratch.old_old_temperature_values[q] *
time_step / old_time_step) :
scratch.old_temperature_values[q]);
(use_bdf2_scheme ? (scratch.old_temperature_grads[q] *
(1 + time_step / old_time_step) -
scratch.old_old_temperature_grads[q] * time_step /
old_time_step) :
scratch.old_temperature_grads[q]);
(use_bdf2_scheme ?
(scratch.old_velocity_values[q] * (1 + time_step / old_time_step) -
scratch.old_old_velocity_values[q] * time_step / old_time_step) :
scratch.old_velocity_values[q]);
(use_bdf2_scheme ?
(scratch.old_strain_rates[q] * (1 + time_step / old_time_step) -
scratch.old_old_strain_rates[q] * time_step / old_time_step) :
scratch.old_strain_rates[q]);
((EquationData::radiogenic_heating * EquationData::density(ext_T) +
2 * EquationData::eta * extrapolated_strain_rate *
extrapolated_strain_rate) /
(EquationData::density(ext_T) * EquationData::specific_heat));
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
data.local_rhs(i) +=
(T_term_for_rhs * scratch.phi_T[i] -
time_step * extrapolated_u * ext_grad_T * scratch.phi_T[i] -
time_step * nu * ext_grad_T * scratch.grad_phi_T[i] +
time_step *
gamma * scratch.phi_T[i]) *
scratch.temperature_fe_values.JxW(q);
if (temperature_constraints.is_inhomogeneously_constrained(
data.local_dof_indices[i]))
{
for (unsigned int j = 0; j < dofs_per_cell; ++j)
data.matrix_for_bc(j, i) +=
(scratch.phi_T[i] * scratch.phi_T[j] *
(use_bdf2_scheme ? ((2 * time_step + old_time_step) /
(time_step + old_time_step)) :
1.) +
scratch.grad_phi_T[i] * scratch.grad_phi_T[j] *
EquationData::kappa * time_step) *
scratch.temperature_fe_values.JxW(q);
}
}
}
}
template <int dim>
void BoussinesqFlowProblem<dim>::copy_local_to_global_temperature_rhs(
const Assembly::CopyData::TemperatureRHS<dim> &data)
{
temperature_constraints.distribute_local_to_global(data.local_rhs,
data.local_dof_indices,
temperature_rhs,
data.matrix_for_bc);
}
void reinit(const Triangulation< dim, spacedim > &tria)
In the function that runs the WorkStream for actually calculating the right hand side, we also generate the final matrix. As mentioned above, it is a sum of the mass matrix and the Laplace matrix, times some time step-dependent weight. This weight is specified by the BDF-2 time integration scheme, see the introduction in step-31. What is new in this tutorial program (in addition to the use of MPI parallelization and the WorkStream class), is that we now precompute the temperature preconditioner as well. The reason is that the setup of the Jacobi preconditioner takes a noticeable time compared to the solver because we usually only need between 10 and 20 iterations for solving the temperature system (this might sound strange, as Jacobi really only consists of a diagonal, but in Trilinos it is derived from more general framework for point relaxation preconditioners which is a bit inefficient). Hence, it is more efficient to precompute the preconditioner, even though the matrix entries may slightly change because the time step might change. This is not too big a problem because we remesh every few time steps (and regenerate the preconditioner then).
template <int dim>
void BoussinesqFlowProblem<dim>::assemble_temperature_system(
const double maximal_velocity)
{
const bool use_bdf2_scheme = (timestep_number != 0);
if (use_bdf2_scheme == true)
{
temperature_matrix.copy_from(temperature_mass_matrix);
temperature_matrix *=
(2 * time_step + old_time_step) / (time_step + old_time_step);
temperature_matrix.add(time_step, temperature_stiffness_matrix);
}
else
{
temperature_matrix.copy_from(temperature_mass_matrix);
temperature_matrix.add(time_step, temperature_stiffness_matrix);
}
if (rebuild_temperature_preconditioner == true)
{
T_preconditioner =
std::make_shared<TrilinosWrappers::PreconditionJacobi>();
T_preconditioner->initialize(temperature_matrix);
rebuild_temperature_preconditioner = false;
}
The next part is computing the right hand side vectors. To do so, we first compute the average temperature \(T_m\) that we use for evaluating the artificial viscosity stabilization through the residual \(E(T) =
(T-T_m)^2\). We do this by defining the midpoint between maximum and minimum temperature as average temperature in the definition of the entropy viscosity. An alternative would be to use the integral average, but the results are not very sensitive to this choice. The rest then only requires calling WorkStream::run again, binding the arguments to the local_assemble_temperature_rhs function that are the same in every call to the correct values:
temperature_rhs = 0;
const QGauss<dim> quadrature_formula(parameters.temperature_degree + 2);
const std::pair<double, double> global_T_range =
get_extrapolated_temperature_range();
const double average_temperature =
0.5 * (global_T_range.first + global_T_range.second);
const double global_entropy_variation =
get_entropy_variation(average_temperature);
using CellFilter =
auto worker =
[this, global_T_range, maximal_velocity, global_entropy_variation](
Assembly::Scratch::TemperatureRHS<dim> & scratch,
Assembly::CopyData::TemperatureRHS<dim> & data) {
this->local_assemble_temperature_rhs(global_T_range,
maximal_velocity,
global_entropy_variation,
cell,
scratch,
data);
};
auto copier = [this](const Assembly::CopyData::TemperatureRHS<dim> &data) {
this->copy_local_to_global_temperature_rhs(data);
};
temperature_dof_handler.begin_active()),
temperature_dof_handler.end()),
worker,
copier,
Assembly::Scratch::TemperatureRHS<dim>(
temperature_fe, stokes_fe, mapping, quadrature_formula),
Assembly::CopyData::TemperatureRHS<dim>(temperature_fe));
}
BoussinesqFlowProblem::solve
This function solves the linear systems in each time step of the Boussinesq problem. First, we work on the Stokes system and then on the temperature system. In essence, it does the same things as the respective function in step-31. However, there are a few changes here.
The first change is related to the way we store our solution: we keep the vectors with locally owned degrees of freedom plus ghost nodes on each MPI node. When we enter a solver which is supposed to perform matrix-vector products with a distributed matrix, this is not the appropriate form, though. There, we will want to have the solution vector to be distributed in the same way as the matrix, i.e. without any ghosts. So what we do first is to generate a distributed vector called distributed_stokes_solution and put only the locally owned dofs into that, which is neatly done by the operator= of the Trilinos vector.
Next, we scale the pressure solution (or rather, the initial guess) for the solver so that it matches with the length scales in the matrices, as discussed in the introduction. We also immediately scale the pressure solution back to the correct units after the solution is completed. We also need to set the pressure values at hanging nodes to zero. This we also did in step-31 in order not to disturb the Schur complement by some vector entries that actually are irrelevant during the solve stage. As a difference to step-31, here we do it only for the locally owned pressure dofs. After solving for the Stokes solution, each processor copies the distributed solution back into the solution vector that also includes ghost elements.
The third and most obvious change is that we have two variants for the Stokes solver: A fast solver that sometimes breaks down, and a robust solver that is slower. This is what we already discussed in the introduction. Here is how we realize it: First, we perform 30 iterations with the fast solver based on the simple preconditioner based on the AMG V-cycle instead of an approximate solve (this is indicated by the false argument to the LinearSolvers::BlockSchurPreconditioner object). If we converge, everything is fine. If we do not converge, the solver control object will throw an exception SolverControl::NoConvergence. Usually, this would abort the program because we don't catch them in our usual solve() functions. This is certainly not what we want to happen here. Rather, we want to switch to the strong solver and continue the solution process with whatever vector we got so far. Hence, we catch the exception with the C++ try/catch mechanism. We then simply go through the same solver sequence again in the catch clause, this time passing the true flag to the preconditioner for the strong solver, signaling an approximate CG solve.
template <int dim>
void BoussinesqFlowProblem<dim>::solve()
{
{
" Solve Stokes system");
pcout << " Solving Stokes system... " << std::flush;
stokes_rhs);
distributed_stokes_solution = stokes_solution;
distributed_stokes_solution.block(1) /= EquationData::pressure_scaling;
const unsigned int
start = (distributed_stokes_solution.block(0).size() +
distributed_stokes_solution.block(1).local_range().first),
end = (distributed_stokes_solution.block(0).size() +
distributed_stokes_solution.block(1).local_range().second);
for (
unsigned int i = start; i <
end; ++i)
if (stokes_constraints.is_constrained(i))
distributed_stokes_solution(i) = 0;
unsigned int n_iterations = 0;
const double solver_tolerance = 1
e-8 * stokes_rhs.l2_norm();
try
{
const LinearSolvers::BlockSchurPreconditioner<
preconditioner(stokes_matrix,
stokes_preconditioner_matrix,
*Mp_preconditioner,
*Amg_preconditioner,
false);
solver_control,
mem,
30));
solver.solve(stokes_matrix,
distributed_stokes_solution,
stokes_rhs,
preconditioner);
n_iterations = solver_control.last_step();
}
{
const LinearSolvers::BlockSchurPreconditioner<
preconditioner(stokes_matrix,
stokes_preconditioner_matrix,
*Mp_preconditioner,
*Amg_preconditioner,
true);
solver_tolerance);
solver_control_refined,
mem,
50));
solver.solve(stokes_matrix,
distributed_stokes_solution,
stokes_rhs,
preconditioner);
n_iterations =
(solver_control.last_step() + solver_control_refined.last_step());
}
stokes_constraints.distribute(distributed_stokes_solution);
distributed_stokes_solution.block(1) *= EquationData::pressure_scaling;
stokes_solution = distributed_stokes_solution;
pcout << n_iterations << " iterations." << std::endl;
}
PETScWrappers::PreconditionBoomerAMG PreconditionAMG
Now let's turn to the temperature part: First, we compute the time step size. We found that we need smaller time steps for 3D than for 2D for the shell geometry. This is because the cells are more distorted in that case (it is the smallest edge length that determines the CFL number). Instead of computing the time step from maximum velocity and minimal mesh size as in step-31, we compute local CFL numbers, i.e., on each cell we compute the maximum velocity times the mesh size, and compute the maximum of them. Hence, we need to choose the factor in front of the time step slightly smaller.
After temperature right hand side assembly, we solve the linear system for temperature (with fully distributed vectors without any ghosts), apply constraints and copy the vector back to one with ghosts.
In the end, we extract the temperature range similarly to step-31 to produce some output (for example in order to help us choose the stabilization constants, as discussed in the introduction). The only difference is that we need to exchange maxima over all processors.
{
" Assemble temperature rhs");
old_time_step = time_step;
const double scaling = (dim == 3 ? 0.25 : 1.0);
time_step = (scaling / (2.1 * dim *
std::sqrt(1. * dim)) /
(parameters.temperature_degree * get_cfl_number()));
const double maximal_velocity = get_maximal_velocity();
pcout << " Maximal velocity: "
<< maximal_velocity * EquationData::year_in_seconds * 100
<< " cm/year" << std::endl;
pcout << " "
<< "Time step: " << time_step / EquationData::year_in_seconds
<< " years" << std::endl;
temperature_solution = old_temperature_solution;
assemble_temperature_system(maximal_velocity);
}
{
" Solve temperature system");
1
e-12 * temperature_rhs.l2_norm());
temperature_rhs);
distributed_temperature_solution = temperature_solution;
cg.solve(temperature_matrix,
distributed_temperature_solution,
temperature_rhs,
*T_preconditioner);
temperature_constraints.distribute(distributed_temperature_solution);
temperature_solution = distributed_temperature_solution;
pcout << " " << solver_control.last_step()
<< " CG iterations for temperature" << std::endl;
double global_temperature[2];
for (unsigned int i =
distributed_temperature_solution.local_range().first;
i < distributed_temperature_solution.local_range().second;
++i)
{
temperature[0] =
std::min<double>(temperature[0],
distributed_temperature_solution(i));
temperature[1] =
std::max<double>(temperature[1],
distributed_temperature_solution(i));
}
temperature[0] *= -1.0;
global_temperature[0] *= -1.0;
pcout << " Temperature range: " << global_temperature[0] << ' '
<< global_temperature[1] << std::endl;
}
}
BoussinesqFlowProblem::output_results
Next comes the function that generates the output. The quantities to output could be introduced manually like we did in step-31. An alternative is to hand this task over to a class PostProcessor that inherits from the class DataPostprocessor, which can be attached to DataOut. This allows us to output derived quantities from the solution, like the friction heating included in this example. It overloads the virtual function DataPostprocessor::evaluate_vector_field(), which is then internally called from DataOut::build_patches(). We have to give it values of the numerical solution, its derivatives, normals to the cell, the actual evaluation points and any additional quantities. This follows the same procedure as discussed in step-29 and other programs.
template <int dim>
class BoussinesqFlowProblem<dim>::Postprocessor
{
public:
Postprocessor(
const unsigned int partition,
const double minimal_pressure);
virtual void evaluate_vector_field(
virtual std::vector<std::string> get_names() const override;
virtual std::vector<
get_data_component_interpretation() const override;
virtual UpdateFlags get_needed_update_flags()
const override;
private:
const double minimal_pressure;
};
template <int dim>
BoussinesqFlowProblem<dim>::Postprocessor::Postprocessor(
const double minimal_pressure)
, minimal_pressure(minimal_pressure)
{}
DataComponentInterpretation
Here we define the names for the variables we want to output. These are the actual solution values for velocity, pressure, and temperature, as well as the friction heating and to each cell the number of the processor that owns it. This allows us to visualize the partitioning of the domain among the processors. Except for the velocity, which is vector-valued, all other quantities are scalar.
template <int dim>
std::vector<std::string>
BoussinesqFlowProblem<dim>::Postprocessor::get_names() const
{
std::vector<std::string> solution_names(dim, "velocity");
solution_names.emplace_back("p");
solution_names.emplace_back("T");
solution_names.emplace_back("friction_heating");
solution_names.emplace_back("partition");
return solution_names;
}
template <int dim>
std::vector<DataComponentInterpretation::DataComponentInterpretation>
BoussinesqFlowProblem<dim>::Postprocessor::get_data_component_interpretation()
const
{
std::vector<DataComponentInterpretation::DataComponentInterpretation>
interpretation(dim,
return interpretation;
}
template <int dim>
BoussinesqFlowProblem<dim>::Postprocessor::get_needed_update_flags() const
{
}
@ component_is_part_of_vector
Now we implement the function that computes the derived quantities. As we also did for the output, we rescale the velocity from its SI units to something more readable, namely cm/year. Next, the pressure is scaled to be between 0 and the maximum pressure. This makes it more easily comparable – in essence making all pressure variables positive or zero. Temperature is taken as is, and the friction heating is computed as \(2 \eta \varepsilon(\mathbf{u}) \cdot \varepsilon(\mathbf{u})\).
The quantities we output here are more for illustration, rather than for actual scientific value. We come back to this briefly in the results section of this program and explain what one may in fact be interested in.
template <int dim>
void BoussinesqFlowProblem<dim>::Postprocessor::evaluate_vector_field(
{
Assert(computed_quantities.size() == n_quadrature_points,
for (unsigned int q = 0; q < n_quadrature_points; ++q)
{
for (
unsigned int d = 0;
d < dim; ++
d)
EquationData::year_in_seconds * 100);
const double pressure =
computed_quantities[q](dim) = pressure;
computed_quantities[q](dim + 1) = temperature;
for (
unsigned int d = 0;
d < dim; ++
d)
computed_quantities[q](dim + 2) =
2 * EquationData::eta * strain_rate * strain_rate;
}
}
constexpr SymmetricTensor< 2, dim, Number > symmetrize(const Tensor< 2, dim, Number > &t)
std::vector<::Vector< double > > solution_values
std::vector< std::vector< Tensor< 1, spacedim > > > solution_gradients
The output_results() function has a similar task to the one in step-31. However, here we are going to demonstrate a different technique on how to merge output from different DoFHandler objects. The way we're going to achieve this recombination is to create a joint DoFHandler that collects both components, the Stokes solution and the temperature solution. This can be nicely done by combining the finite elements from the two systems to form one FESystem, and let this collective system define a new DoFHandler object. To be sure that everything was done correctly, we perform a sanity check that ensures that we got all the dofs from both Stokes and temperature even in the combined system. We then combine the data vectors. Unfortunately, there is no straight-forward relation that tells us how to sort Stokes and temperature vector into the joint vector. The way we can get around this trouble is to rely on the information collected in the FESystem. For each dof on a cell, the joint finite element knows to which equation component (velocity component, pressure, or temperature) it belongs – that's the information we need! So we step through all cells (with iterators into all three DoFHandlers moving in sync), and for each joint cell dof, we read out that component using the FiniteElement::system_to_base_index function (see there for a description of what the various parts of its return value contain). We also need to keep track whether we're on a Stokes dof or a temperature dof, which is contained in joint_fe.system_to_base_index(i).first.first. Eventually, the dof_indices data structures on either of the three systems tell us how the relation between global vector and local dofs looks like on the present cell, which concludes this tedious work. We make sure that each processor only works on the subdomain it owns locally (and not on ghost or artificial cells) when building the joint solution vector. The same will then have to be done in DataOut::build_patches(), but that function does so automatically.
What we end up with is a set of patches that we can write using the functions in DataOutBase in a variety of output formats. Here, we then have to pay attention that what each processor writes is really only its own part of the domain, i.e. we will want to write each processor's contribution into a separate file. This we do by adding an additional number to the filename when we write the solution. This is not really new, we did it similarly in step-40. Note that we write in the compressed format
All the rest of the work is done in the PostProcessor class.
template <int dim>
void BoussinesqFlowProblem<dim>::output_results()
{
joint_dof_handler.distribute_dofs(joint_fe);
Assert(joint_dof_handler.n_dofs() ==
stokes_dof_handler.n_dofs() + temperature_dof_handler.n_dofs(),
joint_solution.
reinit(joint_dof_handler.locally_owned_dofs(),
MPI_COMM_WORLD);
{
std::vector<types::global_dof_index> local_joint_dof_indices(
joint_fe.n_dofs_per_cell());
std::vector<types::global_dof_index> local_stokes_dof_indices(
stokes_fe.n_dofs_per_cell());
std::vector<types::global_dof_index> local_temperature_dof_indices(
temperature_fe.n_dofs_per_cell());
joint_cell = joint_dof_handler.begin_active(),
joint_endc = joint_dof_handler.end(),
stokes_cell = stokes_dof_handler.begin_active(),
temperature_cell = temperature_dof_handler.begin_active();
for (; joint_cell != joint_endc;
++joint_cell, ++stokes_cell, ++temperature_cell)
if (joint_cell->is_locally_owned())
{
joint_cell->get_dof_indices(local_joint_dof_indices);
stokes_cell->get_dof_indices(local_stokes_dof_indices);
temperature_cell->get_dof_indices(local_temperature_dof_indices);
for (unsigned int i = 0; i < joint_fe.n_dofs_per_cell(); ++i)
if (joint_fe.system_to_base_index(i).first.first == 0)
{
Assert(joint_fe.system_to_base_index(i).second <
local_stokes_dof_indices.size(),
joint_solution(local_joint_dof_indices[i]) = stokes_solution(
local_stokes_dof_indices[joint_fe.system_to_base_index(i)
.second]);
}
else
{
Assert(joint_fe.system_to_base_index(i).first.first == 1,
Assert(joint_fe.system_to_base_index(i).second <
local_temperature_dof_indices.size(),
joint_solution(local_joint_dof_indices[i]) =
temperature_solution(
local_temperature_dof_indices
[joint_fe.system_to_base_index(i).second]);
}
}
}
IndexSet locally_relevant_joint_dofs(joint_dof_handler.n_dofs());
locally_relevant_joint_dofs);
locally_relevant_joint_solution.
reinit(locally_relevant_joint_dofs,
MPI_COMM_WORLD);
locally_relevant_joint_solution = joint_solution;
MPI_COMM_WORLD),
stokes_solution.block(1).min());
static int out_index = 0;
"./", "solution", out_index, MPI_COMM_WORLD, 5);
out_index++;
}
void attach_dof_handler(const DoFHandlerType &)
void add_data_vector(const VectorType &data, const std::vector< std::string > &names, const DataVectorType type=type_automatic, const std::vector< DataComponentInterpretation::DataComponentInterpretation > &data_component_interpretation=std::vector< DataComponentInterpretation::DataComponentInterpretation >())
virtual void build_patches(const unsigned int n_subdivisions=0)
std::string write_vtu_with_pvtu_record(const std::string &directory, const std::string &filename_without_extension, const unsigned int counter, const MPI_Comm &mpi_communicator, const unsigned int n_digits_for_counter=numbers::invalid_unsigned_int, const unsigned int n_groups=0) const
void reinit(const Vector &v, const bool omit_zeroing_entries=false, const bool allow_different_maps=false)
void compress(::VectorOperation::values operation)
BoussinesqFlowProblem::refine_mesh
This function isn't really new either. Since the setup_dofs function that we call in the middle has its own timer section, we split timing this function into two sections. It will also allow us to easily identify which of the two is more expensive.
One thing of note, however, is that we only want to compute error indicators on the locally owned subdomain. In order to achieve this, we pass one additional argument to the KellyErrorEstimator::estimate function. Note that the vector for error estimates is resized to the number of active cells present on the current process, which is less than the total number of active cells on all processors (but more than the number of locally owned active cells); each processor only has a few coarse cells around the locally owned ones, as also explained in step-40.
The local error estimates are then handed to a parallel version of GridRefinement (in namespace parallel::distributed::GridRefinement, see also step-40) which looks at the errors and finds the cells that need refinement by comparing the error values across processors. As in step-31, we want to limit the maximum grid level. So in case some cells have been marked that are already at the finest level, we simply clear the refine flags.
template <int dim>
void
BoussinesqFlowProblem<dim>::refine_mesh(const unsigned int max_grid_level)
{
temperature_trans(temperature_dof_handler);
stokes_trans(stokes_dof_handler);
{
"Refine mesh structure, part 1");
temperature_dof_handler,
temperature_solution,
estimated_error_per_cell,
nullptr,
0,
++cell)
cell->clear_refine_flag();
static void estimate(const Mapping< dim, spacedim > &mapping, const DoFHandler< dim, spacedim > &dof, const Quadrature< dim - 1 > &quadrature, const std::map< types::boundary_id, const Function< spacedim, typename InputVector::value_type > * > &neumann_bc, const InputVector &solution, Vector< float > &error, const ComponentMask &component_mask=ComponentMask(), const Function< spacedim > *coefficients=nullptr, const unsigned int n_threads=numbers::invalid_unsigned_int, const types::subdomain_id subdomain_id=numbers::invalid_subdomain_id, const types::material_id material_id=numbers::invalid_material_id, const Strategy strategy=cell_diameter_over_24)
void refine_and_coarsen_fixed_fraction(parallel::distributed::Triangulation< dim, spacedim > &tria, const ::Vector< Number > &criteria, const double top_fraction_of_error, const double bottom_fraction_of_error, const VectorTools::NormType norm_type=VectorTools::NormType::L1_norm)
With all flags marked as necessary, we can then tell the parallel::distributed::SolutionTransfer objects to get ready to transfer data from one mesh to the next, which they will do when notified by Triangulation as part of the execute_coarsening_and_refinement() call. The syntax is similar to the non-parallel solution transfer (with the exception that here a pointer to the vector entries is enough). The remainder of the function further down below is then concerned with setting up the data structures again after mesh refinement and restoring the solution vectors on the new mesh.
std::vector<const TrilinosWrappers::MPI::Vector *> x_temperature(2);
x_temperature[0] = &temperature_solution;
x_temperature[1] = &old_temperature_solution;
std::vector<const TrilinosWrappers::MPI::BlockVector *> x_stokes(2);
x_stokes[0] = &stokes_solution;
x_stokes[1] = &old_stokes_solution;
temperature_trans.prepare_for_coarsening_and_refinement(x_temperature);
stokes_trans.prepare_for_coarsening_and_refinement(x_stokes);
}
setup_dofs();
{
"Refine mesh structure, part 2");
{
std::vector<TrilinosWrappers::MPI::Vector *> tmp(2);
tmp[0] = &(distributed_temp1);
tmp[1] = &(distributed_temp2);
temperature_trans.interpolate(tmp);
enforce constraints to make the interpolated solution conforming on the new mesh:
temperature_constraints.distribute(distributed_temp1);
temperature_constraints.distribute(distributed_temp2);
temperature_solution = distributed_temp1;
old_temperature_solution = distributed_temp2;
}
{
std::vector<TrilinosWrappers::MPI::BlockVector *> stokes_tmp(2);
stokes_tmp[0] = &(distributed_stokes);
stokes_tmp[1] = &(old_distributed_stokes);
stokes_trans.interpolate(stokes_tmp);
enforce constraints to make the interpolated solution conforming on the new mesh:
stokes_constraints.distribute(distributed_stokes);
stokes_constraints.distribute(old_distributed_stokes);
stokes_solution = distributed_stokes;
old_stokes_solution = old_distributed_stokes;
}
}
}
BoussinesqFlowProblem::run
This is the final and controlling function in this class. It, in fact, runs the entire rest of the program and is, once more, very similar to step-31. The only substantial difference is that we use a different mesh now (a GridGenerator::hyper_shell instead of a simple cube geometry).
template <int dim>
{
EquationData::R0,
EquationData::R1,
(dim == 3) ? 96 : 12,
true);
triangulation.refine_global(parameters.initial_global_refinement);
setup_dofs();
unsigned int pre_refinement_step = 0;
start_time_iteration:
{
temperature_dof_handler.locally_owned_dofs());
void hyper_shell(Triangulation< dim > &tria, const Point< dim > ¢er, const double inner_radius, const double outer_radius, const unsigned int n_cells=0, bool colorize=false)
VectorTools::project supports parallel vector classes with most standard finite elements via deal.II's own native MatrixFree framework: since we use standard Lagrange elements of moderate order this function works well here.
temperature_constraints,
EquationData::TemperatureInitialValues<dim>(),
solution);
Having so computed the current temperature field, let us set the member variable that holds the temperature nodes. Strictly speaking, we really only need to set old_temperature_solution since the first thing we will do is to compute the Stokes solution that only requires the previous time step's temperature field. That said, nothing good can come from not initializing the other vectors as well (especially since it's a relatively cheap operation and we only have to do it once at the beginning of the program) if we ever want to extend our numerical method or physical model, and so we initialize old_temperature_solution and old_old_temperature_solution as well. The assignment makes sure that the vectors on the left hand side (which where initialized to contain ghost elements as well) also get the correct ghost elements. In other words, the assignment here requires communication between processors:
temperature_solution = solution;
old_temperature_solution = solution;
old_old_temperature_solution = solution;
}
timestep_number = 0;
time_step = old_time_step = 0;
double time = 0;
do
{
pcout << "Timestep " << timestep_number
<< ": t=" << time / EquationData::year_in_seconds << " years"
<< std::endl;
assemble_stokes_system();
build_stokes_preconditioner();
assemble_temperature_matrix();
solve();
pcout << std::endl;
if ((timestep_number == 0) &&
(pre_refinement_step < parameters.initial_adaptive_refinement))
{
refine_mesh(parameters.initial_global_refinement +
parameters.initial_adaptive_refinement);
++pre_refinement_step;
goto start_time_iteration;
}
else if ((timestep_number > 0) &&
(timestep_number % parameters.adaptive_refinement_interval ==
0))
refine_mesh(parameters.initial_global_refinement +
parameters.initial_adaptive_refinement);
if ((parameters.generate_graphical_output == true) &&
(timestep_number % parameters.graphical_output_interval == 0))
output_results();
In order to speed up linear solvers, we extrapolate the solutions from the old time levels to the new one. This gives a very good initial guess, cutting the number of iterations needed in solvers by more than one half. We do not need to extrapolate in the last iteration, so if we reached the final time, we stop here.
As the last thing during a time step (before actually bumping up the number of the time step), we check whether the current time step number is divisible by 100, and if so we let the computing timer print a summary of CPU times spent so far.
if (time > parameters.end_time * EquationData::year_in_seconds)
break;
old_old_stokes_solution = old_stokes_solution;
old_stokes_solution = stokes_solution;
old_old_temperature_solution = old_temperature_solution;
old_temperature_solution = temperature_solution;
if (old_time_step > 0)
{
Trilinos sadd does not like ghost vectors even as input. Copy into distributed vectors for now:
{
distr_solution = stokes_solution;
distr_old_solution = old_old_stokes_solution;
distr_solution.
sadd(1. + time_step / old_time_step,
-time_step / old_time_step,
distr_old_solution);
stokes_solution = distr_solution;
}
{
distr_solution = temperature_solution;
distr_old_solution = old_old_temperature_solution;
distr_solution.sadd(1. + time_step / old_time_step,
-time_step / old_time_step,
distr_old_solution);
temperature_solution = distr_solution;
}
}
if ((timestep_number > 0) && (timestep_number % 100 == 0))
computing_timer.print_summary();
time += time_step;
++timestep_number;
}
while (true);
void sadd(const value_type s, const BlockVectorBase &V)
If we are generating graphical output, do so also for the last time step unless we had just done so before we left the do-while loop
if ((parameters.generate_graphical_output == true) &&
!((timestep_number - 1) % parameters.graphical_output_interval == 0))
output_results();
}
}
The main function
The main function is short as usual and very similar to the one in step-31. Since we use a parameter file which is specified as an argument in the command line, we have to read it in here and pass it on to the Parameters class for parsing. If no filename is given in the command line, we simply use the step-32.prm file which is distributed together with the program.
Because 3d computations are simply very slow unless you throw a lot of processors at them, the program defaults to 2d. You can get the 3d version by changing the constant dimension below to 3.
int main(int argc, char *argv[])
{
try
{
using namespace Step32;
std::string parameter_filename;
if (argc >= 2)
parameter_filename = argv[1];
else
parameter_filename = "step-32.prm";
const int dim = 2;
BoussinesqFlowProblem<dim>::Parameters parameters(parameter_filename);
BoussinesqFlowProblem<dim> flow_problem(parameters);
flow_problem.run();
}
catch (std::exception &exc)
{
std::cerr << std::endl
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Exception on processing: " << std::endl
<< exc.what() << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
catch (...)
{
std::cerr << std::endl
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Unknown exception!" << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
return 0;
}
static const unsigned int invalid_unsigned_int
Results
When run, the program simulates convection in 3d in much the same way as step-31 did, though with an entirely different testcase.
Comparison of results with step-31
Before we go to this testcase, however, let us show a few results from a slightly earlier version of this program that was solving exactly the testcase we used in step-31, just that we now solve it in parallel and with much higher resolution. We show these results mainly for comparison.
Here are two images that show this higher resolution if we choose a 3d computation in main() and if we set initial_refinement=3 and n_pre_refinement_steps=4. At the time steps shown, the meshes had around 72,000 and 236,000 cells, for a total of 2,680,000 and 8,250,000 degrees of freedom, respectively, more than an order of magnitude more than we had available in step-31:
The computation was done on a subset of 50 processors of the Brazos cluster at Texas A&M University.
Results for a 2d circular shell testcase
Next, we will run step-32 with the parameter file in the directory with one change: we increase the final time to 1e9. Here we are using 16 processors. The command to launch is (note that step-32.prm is the default):
$ mpirun -np 16 ./step-32
Note that running a job on a cluster typically requires going through a job scheduler, which we won't discuss here. The output will look roughly like this:
$ mpirun -np 16 ./step-32
Number of active cells: 12,288 (on 6 levels)
Number of degrees of freedom: 186,624 (99,840+36,864+49,920)
Timestep 0: t=0 years
Rebuilding Stokes preconditioner...
Solving Stokes system... 41 iterations.
Maximal velocity: 60.4935 cm/year
Time step: 18166.9 years
17 CG iterations for temperature
Temperature range: 973 4273.16
Number of active cells: 15,921 (on 7 levels)
Number of degrees of freedom: 252,723 (136,640+47,763+68,320)
Timestep 0: t=0 years
Rebuilding Stokes preconditioner...
Solving Stokes system... 50 iterations.
Maximal velocity: 60.3223 cm/year
Time step: 10557.6 years
19 CG iterations for temperature
Temperature range: 973 4273.16
Number of active cells: 19,926 (on 8 levels)
Number of degrees of freedom: 321,246 (174,312+59,778+87,156)
Timestep 0: t=0 years
Rebuilding Stokes preconditioner...
Solving Stokes system... 50 iterations.
Maximal velocity: 57.8396 cm/year
Time step: 5453.78 years
18 CG iterations for temperature
Temperature range: 973 4273.16
Timestep 1: t=5453.78 years
Solving Stokes system... 49 iterations.
Maximal velocity: 59.0231 cm/year
Time step: 5345.86 years
18 CG iterations for temperature
Temperature range: 973 4273.16
Timestep 2: t=10799.6 years
Solving Stokes system... 24 iterations.
Maximal velocity: 60.2139 cm/year
Time step: 5241.51 years
17 CG iterations for temperature
Temperature range: 973 4273.16
[...]
Timestep 100: t=272151 years
Solving Stokes system... 21 iterations.
Maximal velocity: 161.546 cm/year
Time step: 1672.96 years
17 CG iterations for temperature
Temperature range: 973 4282.57
Number of active cells: 56,085 (on 8 levels)
Number of degrees of freedom: 903,408 (490,102+168,255+245,051)
+---------------------------------------------+------------+------------+
| Total wallclock time elapsed since start | 115s | |
| | | |
| Section | no. calls | wall time | % of total |
+---------------------------------+-----------+------------+------------+
| Assemble Stokes system | 103 | 2.82s | 2.5% |
| Assemble temperature matrices | 12 | 0.452s | 0.39% |
| Assemble temperature rhs | 103 | 11.5s | 10% |
| Build Stokes preconditioner | 12 | 2.09s | 1.8% |
| Solve Stokes system | 103 | 90.4s | 79% |
| Solve temperature system | 103 | 1.53s | 1.3% |
| Postprocessing | 3 | 0.532s | 0.46% |
| Refine mesh structure, part 1 | 12 | 0.93s | 0.81% |
| Refine mesh structure, part 2 | 12 | 0.384s | 0.33% |
| Setup dof systems | 13 | 2.96s | 2.6% |
+---------------------------------+-----------+------------+------------+
[...]
+---------------------------------------------+------------+------------+
| Total wallclock time elapsed since start | 9.14e+04s | |
| | | |
| Section | no. calls | wall time | % of total |
+---------------------------------+-----------+------------+------------+
| Assemble Stokes system | 47045 | 2.05e+03s | 2.2% |
| Assemble temperature matrices | 4707 | 310s | 0.34% |
| Assemble temperature rhs | 47045 | 8.7e+03s | 9.5% |
| Build Stokes preconditioner | 4707 | 1.48e+03s | 1.6% |
| Solve Stokes system | 47045 | 7.34e+04s | 80% |
| Solve temperature system | 47045 | 1.46e+03s | 1.6% |
| Postprocessing | 1883 | 222s | 0.24% |
| Refine mesh structure, part 1 | 4706 | 641s | 0.7% |
| Refine mesh structure, part 2 | 4706 | 259s | 0.28% |
| Setup dof systems | 4707 | 1.86e+03s | 2% |
+---------------------------------+-----------+------------+------------+
The simulation terminates when the time reaches the 1 billion years selected in the input file. You can extrapolate from this how long a simulation would take for a different final time (the time step size ultimately settles on somewhere around 20,000 years, so computing for two billion years will take 100,000 time steps, give or take 20%). As can be seen here, we spend most of the compute time in assembling linear systems and — above all — in solving Stokes systems.
To demonstrate the output we show the output from every 1250th time step here:
The last two images show the grid as well as the partitioning of the mesh for the same computation with 16 subdomains and 16 processors. The full dynamics of this simulation are really only visible by looking at an animation, for example the one shown on this site. This image is well worth watching due to its artistic quality and entrancing depiction of the evolution of the magma plumes.
If you watch the movie, you'll see that the convection pattern goes through several stages: First, it gets rid of the instable temperature layering with the hot material overlain by the dense cold material. After this great driver is removed and we have a sort of stable situation, a few blobs start to separate from the hot boundary layer at the inner ring and rise up, with a few cold fingers also dropping down from the outer boundary layer. During this phase, the solution remains mostly symmetric, reflecting the 12-fold symmetry of the original mesh. In a final phase, the fluid enters vigorous chaotic stirring in which all symmetries are lost. This is a pattern that then continues to dominate flow.
These different phases can also be identified if we look at the maximal velocity as a function of time in the simulation:
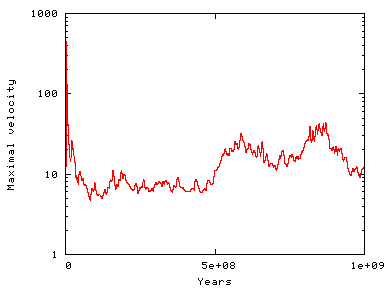
Here, the velocity (shown in centimeters per year) becomes very large, to the order of several meters per year) at the beginning when the temperature layering is instable. It then calms down to relatively small values before picking up again in the chaotic stirring regime. There, it remains in the range of 10-40 centimeters per year, quite within the physically expected region.
Results for a 3d spherical shell testcase
3d computations are very expensive computationally. Furthermore, as seen above, interesting behavior only starts after quite a long time requiring more CPU hours than is available on a typical cluster. Consequently, rather than showing a complete simulation here, let us simply show a couple of pictures we have obtained using the successor to this program, called ASPECT (short for Advanced Solver for Problems in Earth's ConvecTion), that is being developed independently of deal.II and that already incorporates some of the extensions discussed below. The following two pictures show isocontours of the temperature and the partition of the domain (along with the mesh) onto 512 processors:

Possibilities for extensions
There are many directions in which this program could be extended. As mentioned at the end of the introduction, most of these are under active development in the ASPECT (short for Advanced Solver for Problems in Earth's ConvecTion) code at the time this tutorial program is being finished. Specifically, the following are certainly topics that one should address to make the program more useful:
-
Adiabatic heating/cooling: The temperature field we get in our simulations after a while is mostly constant with boundary layers at the inner and outer boundary, and streamers of cold and hot material mixing everything. Yet, this doesn't match our expectation that things closer to the earth core should be hotter than closer to the surface. The reason is that the energy equation we have used does not include a term that describes adiabatic cooling and heating: rock, like gas, heats up as you compress it. Consequently, material that rises up cools adiabatically, and cold material that sinks down heats adiabatically. The correct temperature equation would therefore look somewhat like this:
\begin{eqnarray*}
\frac{D T}{Dt}
-
\nabla \cdot \kappa \nabla T &=& \gamma + \tau\frac{Dp}{Dt},
\end{eqnarray*}
or, expanding the advected derivative \(\frac{D}{Dt} =
\frac{\partial}{\partial t} + \mathbf u \cdot \nabla\):
\begin{eqnarray*}
\frac{\partial T}{\partial t}
+
{\mathbf u} \cdot \nabla T
-
\nabla \cdot \kappa \nabla T &=& \gamma +
\tau\left\{\frac{\partial
p}{\partial t} + \mathbf u \cdot \nabla p \right\}.
\end{eqnarray*}
In other words, as pressure increases in a rock volume ( \(\frac{Dp}{Dt}>0\)) we get an additional heat source, and vice versa.
The time derivative of the pressure is a bit awkward to implement. If necessary, one could approximate using the fact outlined in the introduction that the pressure can be decomposed into a dynamic component due to temperature differences and the resulting flow, and a static component that results solely from the static pressure of the overlying rock. Since the latter is much bigger, one may approximate \(p\approx p_{\text{static}}=-\rho_{\text{ref}}
[1+\beta T_{\text{ref}}] \varphi\), and consequently \(\frac{Dp}{Dt} \approx \left\{- \mathbf u \cdot \nabla \rho_{\text{ref}}
[1+\beta T_{\text{ref}}]\varphi\right\} = \rho_{\text{ref}}
[1+\beta T_{\text{ref}}] \mathbf u \cdot \mathbf g\). In other words, if the fluid is moving in the direction of gravity (downward) it will be compressed and because in that case \(\mathbf u
\cdot \mathbf g > 0\) we get a positive heat source. Conversely, the fluid will cool down if it moves against the direction of gravity.
-
Compressibility: As already hinted at in the temperature model above, mantle rocks are not incompressible. Rather, given the enormous pressures in the earth mantle (at the core-mantle boundary, the pressure is approximately 140 GPa, equivalent to 1,400,000 times atmospheric pressure), rock actually does compress to something around 1.5 times the density it would have at surface pressure. Modeling this presents any number of difficulties. Primarily, the mass conservation equation is no longer \(\textrm{div}\;\mathbf u=0\) but should read \(\textrm{div}(\rho\mathbf u)=0\) where the density \(\rho\) is now no longer spatially constant but depends on temperature and pressure. A consequence is that the model is now no longer linear; a linearized version of the Stokes equation is also no longer symmetric requiring us to rethink preconditioners and, possibly, even the discretization. We won't go into detail here as to how this can be resolved.
-
Nonlinear material models: As already hinted at in various places, material parameters such as the density, the viscosity, and the various thermal parameters are not constant throughout the earth mantle. Rather, they nonlinearly depend on the pressure and temperature, and in the case of the viscosity on the strain rate \(\varepsilon(\mathbf u)\). For complicated models, the only way to solve such models accurately may be to actually iterate this dependence out in each time step, rather than simply freezing coefficients at values extrapolated from the previous time step(s).
-
Checkpoint/restart: Running this program in 2d on a number of processors allows solving realistic models in a day or two. However, in 3d, compute times are so large that one runs into two typical problems: (i) On most compute clusters, the queuing system limits run times for individual jobs are to 2 or 3 days; (ii) losing the results of a computation due to hardware failures, misconfigurations, or power outages is a shame when running on hundreds of processors for a couple of days. Both of these problems can be addressed by periodically saving the state of the program and, if necessary, restarting the program at this point. This technique is commonly called checkpoint/restart and it requires that the entire state of the program is written to a permanent storage location (e.g. a hard drive). Given the complexity of the data structures of this program, this is not entirely trivial (it may also involve writing gigabytes or more of data), but it can be made easier by realizing that one can save the state between two time steps where it essentially only consists of the mesh and solution vectors; during restart one would then first re-enumerate degrees of freedom in the same way as done before and then re-assemble matrices. Nevertheless, given the distributed nature of the data structures involved here, saving and restoring the state of a program is not trivial. An additional complexity is introduced by the fact that one may want to change the number of processors between runs, for example because one may wish to continue computing on a mesh that is finer than the one used to precompute a starting temperature field at an intermediate time.
-
Predictive postprocessing: The point of computations like this is not simply to solve the equations. Rather, it is typically the exploration of different physical models and their comparison with things that we can measure at the earth surface, in order to find which models are realistic and which are contradicted by reality. To this end, we need to compute quantities from our solution vectors that are related to what we can observe. Among these are, for example, heatfluxes at the surface of the earth, as well as seismic velocities throughout the mantle as these affect earthquake waves that are recorded by seismographs.
-
Better refinement criteria: As can be seen above for the 3d case, the mesh in 3d is primarily refined along the inner boundary. This is because the boundary layer there is stronger than any other transition in the domain, leading us to refine there almost exclusively and basically not at all following the plumes. One certainly needs better refinement criteria to track the parts of the solution we are really interested in better than the criterion used here, namely the KellyErrorEstimator applied to the temperature, is able to.
There are many other ways to extend the current program. However, rather than discussing them here, let us point to the much larger open source code ASPECT (see https://aspect.geodynamics.org/ ) that constitutes the further development of step-32 and that already includes many such possible extensions.
The plain program
#include <fstream>
#include <iostream>
#include <limits>
#include <locale>
#include <string>
namespace Step32
{
namespace EquationData
{
constexpr double eta = 1e21;
constexpr double kappa = 1
e-6;
constexpr double reference_density = 3300;
constexpr double reference_temperature = 293;
constexpr double expansion_coefficient = 2
e-5;
constexpr double specific_heat = 1250;
constexpr double radiogenic_heating = 7.4e-12;
constexpr double R0 = 6371000. - 2890000.;
constexpr double R1 = 6371000. - 35000.;
constexpr double T0 = 4000 + 273;
constexpr double T1 = 700 + 273;
double density(const double temperature)
{
return (
reference_density *
(1 - expansion_coefficient * (temperature - reference_temperature)));
}
template <int dim>
{
const double r = p.
norm();
return -(1.245e-6 * r + 7.714e13 / r / r) * p / r;
}
template <int dim>
class TemperatureInitialValues :
public Function<dim>
{
public:
TemperatureInitialValues()
{}
const unsigned int component = 0) const override;
};
template <int dim>
double TemperatureInitialValues<dim>::value(
const Point<dim> &p,
const unsigned int) const
{
const double r = p.
norm();
const double h = R1 - R0;
const double s = (r - R0) / h;
const double q =
const double tau = s + 0.2 * s * (1 - s) *
std::sin(6 * phi) * q;
return T0 * (1.0 - tau) + T1 * tau;
}
template <int dim>
void
TemperatureInitialValues<dim>::vector_value(
const Point<dim> &p,
{
for (unsigned int c = 0; c < this->n_components; ++c)
values(c) = TemperatureInitialValues<dim>::value(p, c);
}
constexpr double pressure_scaling = eta / 10000;
const double year_in_seconds = 60 * 60 * 24 * 365.2425;
}
namespace LinearSolvers
{
template <class PreconditionerTypeA, class PreconditionerTypeMp>
{
public:
const PreconditionerTypeMp &Mppreconditioner,
const PreconditionerTypeA & Apreconditioner,
const bool do_solve_A)
: stokes_matrix(&S)
, stokes_preconditioner_matrix(&Spre)
, mp_preconditioner(Mppreconditioner)
, a_preconditioner(Apreconditioner)
, do_solve_A(do_solve_A)
{}
{
{
solver.solve(stokes_preconditioner_matrix->block(1, 1),
mp_preconditioner);
}
{
stokes_matrix->block(0, 1).vmult(utmp, dst.
block(1));
utmp *= -1.0;
}
if (do_solve_A == true)
{
solver.solve(stokes_matrix->block(0, 0),
utmp,
a_preconditioner);
}
else
a_preconditioner.vmult(dst.
block(0), utmp);
}
private:
stokes_matrix;
stokes_preconditioner_matrix;
const PreconditionerTypeMp &mp_preconditioner;
const PreconditionerTypeA & a_preconditioner;
const bool do_solve_A;
};
}
namespace Assembly
{
namespace Scratch
{
template <int dim>
struct StokesPreconditioner
{
StokesPreconditioner(const StokesPreconditioner &data);
std::vector<Tensor<2, dim>> grad_phi_u;
std::vector<double> phi_p;
};
template <int dim>
StokesPreconditioner<dim>::StokesPreconditioner(
: stokes_fe_values(mapping, stokes_fe, stokes_quadrature, update_flags)
, grad_phi_u(stokes_fe.n_dofs_per_cell())
, phi_p(stokes_fe.n_dofs_per_cell())
{}
template <int dim>
StokesPreconditioner<dim>::StokesPreconditioner(
const StokesPreconditioner &scratch)
: stokes_fe_values(scratch.stokes_fe_values.get_mapping(),
scratch.stokes_fe_values.get_fe(),
scratch.stokes_fe_values.get_quadrature(),
scratch.stokes_fe_values.get_update_flags())
, grad_phi_u(scratch.grad_phi_u)
, phi_p(scratch.phi_p)
{}
template <int dim>
struct StokesSystem : public StokesPreconditioner<dim>
{
StokesSystem(const StokesSystem<dim> &data);
std::vector<Tensor<1, dim>> phi_u;
std::vector<SymmetricTensor<2, dim>> grads_phi_u;
std::vector<double> div_phi_u;
std::vector<double> old_temperature_values;
};
template <int dim>
StokesSystem<dim>::StokesSystem(
: StokesPreconditioner<dim>(stokes_fe,
stokes_quadrature,
mapping,
stokes_update_flags)
, temperature_fe_values(mapping,
temperature_fe,
stokes_quadrature,
temperature_update_flags)
, phi_u(stokes_fe.n_dofs_per_cell())
, grads_phi_u(stokes_fe.n_dofs_per_cell())
, div_phi_u(stokes_fe.n_dofs_per_cell())
, old_temperature_values(stokes_quadrature.size())
{}
template <int dim>
StokesSystem<dim>::StokesSystem(const StokesSystem<dim> &scratch)
: StokesPreconditioner<dim>(scratch)
, temperature_fe_values(
scratch.temperature_fe_values.get_mapping(),
scratch.temperature_fe_values.get_fe(),
scratch.temperature_fe_values.get_quadrature(),
scratch.temperature_fe_values.get_update_flags())
, phi_u(scratch.phi_u)
, grads_phi_u(scratch.grads_phi_u)
, div_phi_u(scratch.div_phi_u)
, old_temperature_values(scratch.old_temperature_values)
{}
template <int dim>
struct TemperatureMatrix
{
TemperatureMatrix(const TemperatureMatrix &data);
std::vector<double> phi_T;
std::vector<Tensor<1, dim>> grad_phi_T;
};
template <int dim>
TemperatureMatrix<dim>::TemperatureMatrix(
: temperature_fe_values(mapping,
temperature_fe,
temperature_quadrature,
, phi_T(temperature_fe.n_dofs_per_cell())
, grad_phi_T(temperature_fe.n_dofs_per_cell())
{}
template <int dim>
TemperatureMatrix<dim>::TemperatureMatrix(
const TemperatureMatrix &scratch)
: temperature_fe_values(
scratch.temperature_fe_values.get_mapping(),
scratch.temperature_fe_values.get_fe(),
scratch.temperature_fe_values.get_quadrature(),
scratch.temperature_fe_values.get_update_flags())
, phi_T(scratch.phi_T)
, grad_phi_T(scratch.grad_phi_T)
{}
template <int dim>
struct TemperatureRHS
{
TemperatureRHS(const TemperatureRHS &data);
std::vector<double> phi_T;
std::vector<Tensor<1, dim>> grad_phi_T;
std::vector<Tensor<1, dim>> old_velocity_values;
std::vector<Tensor<1, dim>> old_old_velocity_values;
std::vector<SymmetricTensor<2, dim>> old_strain_rates;
std::vector<SymmetricTensor<2, dim>> old_old_strain_rates;
std::vector<double> old_temperature_values;
std::vector<double> old_old_temperature_values;
std::vector<Tensor<1, dim>> old_temperature_grads;
std::vector<Tensor<1, dim>> old_old_temperature_grads;
std::vector<double> old_temperature_laplacians;
std::vector<double> old_old_temperature_laplacians;
};
template <int dim>
TemperatureRHS<dim>::TemperatureRHS(
: temperature_fe_values(mapping,
temperature_fe,
quadrature,
, stokes_fe_values(mapping,
stokes_fe,
quadrature,
, phi_T(temperature_fe.n_dofs_per_cell())
, grad_phi_T(temperature_fe.n_dofs_per_cell())
,
old_velocity_values(quadrature.size())
, old_old_velocity_values(quadrature.size())
, old_strain_rates(quadrature.size())
, old_old_strain_rates(quadrature.size())
,
old_temperature_values(quadrature.size())
, old_old_temperature_values(quadrature.size())
, old_temperature_grads(quadrature.size())
, old_old_temperature_grads(quadrature.size())
, old_temperature_laplacians(quadrature.size())
, old_old_temperature_laplacians(quadrature.size())
{}
template <int dim>
TemperatureRHS<dim>::TemperatureRHS(const TemperatureRHS &scratch)
: temperature_fe_values(
scratch.temperature_fe_values.get_mapping(),
scratch.temperature_fe_values.get_fe(),
scratch.temperature_fe_values.get_quadrature(),
scratch.temperature_fe_values.get_update_flags())
, stokes_fe_values(scratch.stokes_fe_values.get_mapping(),
scratch.stokes_fe_values.get_fe(),
scratch.stokes_fe_values.get_quadrature(),
scratch.stokes_fe_values.get_update_flags())
, phi_T(scratch.phi_T)
, grad_phi_T(scratch.grad_phi_T)
,
old_velocity_values(scratch.old_velocity_values)
, old_old_velocity_values(scratch.old_old_velocity_values)
, old_strain_rates(scratch.old_strain_rates)
, old_old_strain_rates(scratch.old_old_strain_rates)
,
old_temperature_values(scratch.old_temperature_values)
, old_old_temperature_values(scratch.old_old_temperature_values)
, old_temperature_grads(scratch.old_temperature_grads)
, old_old_temperature_grads(scratch.old_old_temperature_grads)
, old_temperature_laplacians(scratch.old_temperature_laplacians)
, old_old_temperature_laplacians(scratch.old_old_temperature_laplacians)
{}
}
namespace CopyData
{
template <int dim>
struct StokesPreconditioner
{
StokesPreconditioner(const StokesPreconditioner &data);
StokesPreconditioner &operator=(const StokesPreconditioner &) = default;
std::vector<types::global_dof_index> local_dof_indices;
};
template <int dim>
StokesPreconditioner<dim>::StokesPreconditioner(
: local_matrix(stokes_fe.n_dofs_per_cell(), stokes_fe.n_dofs_per_cell())
, local_dof_indices(stokes_fe.n_dofs_per_cell())
{}
template <int dim>
StokesPreconditioner<dim>::StokesPreconditioner(
const StokesPreconditioner &data)
: local_matrix(data.local_matrix)
, local_dof_indices(data.local_dof_indices)
{}
template <int dim>
struct StokesSystem : public StokesPreconditioner<dim>
{
};
template <int dim>
: StokesPreconditioner<dim>(stokes_fe)
, local_rhs(stokes_fe.n_dofs_per_cell())
{}
template <int dim>
struct TemperatureMatrix
{
std::vector<types::global_dof_index> local_dof_indices;
};
template <int dim>
TemperatureMatrix<dim>::TemperatureMatrix(
: local_mass_matrix(temperature_fe.n_dofs_per_cell(),
temperature_fe.n_dofs_per_cell())
, local_stiffness_matrix(temperature_fe.n_dofs_per_cell(),
temperature_fe.n_dofs_per_cell())
, local_dof_indices(temperature_fe.n_dofs_per_cell())
{}
template <int dim>
struct TemperatureRHS
{
std::vector<types::global_dof_index> local_dof_indices;
};
template <int dim>
TemperatureRHS<dim>::TemperatureRHS(
: local_rhs(temperature_fe.n_dofs_per_cell())
, local_dof_indices(temperature_fe.n_dofs_per_cell())
, matrix_for_bc(temperature_fe.n_dofs_per_cell(),
temperature_fe.n_dofs_per_cell())
{}
}
}
template <int dim>
class BoussinesqFlowProblem
{
public:
struct Parameters;
BoussinesqFlowProblem(Parameters ¶meters);
private:
void setup_dofs();
void assemble_stokes_preconditioner();
void build_stokes_preconditioner();
void assemble_stokes_system();
void assemble_temperature_matrix();
void assemble_temperature_system(const double maximal_velocity);
double get_maximal_velocity() const;
double get_cfl_number() const;
double get_entropy_variation(const double average_temperature) const;
std::pair<double, double> get_extrapolated_temperature_range() const;
void solve();
void output_results();
void refine_mesh(const unsigned int max_grid_level);
double compute_viscosity(
const std::vector<double> & old_temperature,
const std::vector<double> & old_old_temperature,
const std::vector<double> & old_temperature_laplacians,
const std::vector<double> & old_old_temperature_laplacians,
const double global_u_infty,
const double global_T_variation,
const double average_temperature,
const double global_entropy_variation,
const double cell_diameter) const;
public:
struct Parameters
{
Parameters(const std::string ¶meter_filename);
double end_time;
unsigned int initial_global_refinement;
unsigned int initial_adaptive_refinement;
bool generate_graphical_output;
unsigned int graphical_output_interval;
unsigned int adaptive_refinement_interval;
double stabilization_alpha;
double stabilization_c_R;
double stabilization_beta;
unsigned int stokes_velocity_degree;
bool use_locally_conservative_discretization;
unsigned int temperature_degree;
};
private:
Parameters ¶meters;
double global_Omega_diameter;
double time_step;
double old_time_step;
unsigned int timestep_number;
std::shared_ptr<TrilinosWrappers::PreconditionAMG> Amg_preconditioner;
std::shared_ptr<TrilinosWrappers::PreconditionJacobi> Mp_preconditioner;
std::shared_ptr<TrilinosWrappers::PreconditionJacobi> T_preconditioner;
bool rebuild_stokes_matrix;
bool rebuild_stokes_preconditioner;
bool rebuild_temperature_matrices;
bool rebuild_temperature_preconditioner;
void setup_stokes_matrix(
const std::vector<IndexSet> &stokes_partitioning,
const std::vector<IndexSet> &stokes_relevant_partitioning);
void setup_stokes_preconditioner(
const std::vector<IndexSet> &stokes_partitioning,
const std::vector<IndexSet> &stokes_relevant_partitioning);
void setup_temperature_matrices(
const IndexSet &temperature_partitioning,
const IndexSet &temperature_relevant_partitioning);
void local_assemble_stokes_preconditioner(
Assembly::Scratch::StokesPreconditioner<dim> & scratch,
Assembly::CopyData::StokesPreconditioner<dim> & data);
void copy_local_to_global_stokes_preconditioner(
const Assembly::CopyData::StokesPreconditioner<dim> &data);
void local_assemble_stokes_system(
Assembly::Scratch::StokesSystem<dim> & scratch,
Assembly::CopyData::StokesSystem<dim> & data);
void copy_local_to_global_stokes_system(
const Assembly::CopyData::StokesSystem<dim> &data);
void local_assemble_temperature_matrix(
Assembly::Scratch::TemperatureMatrix<dim> & scratch,
Assembly::CopyData::TemperatureMatrix<dim> & data);
void copy_local_to_global_temperature_matrix(
const Assembly::CopyData::TemperatureMatrix<dim> &data);
void local_assemble_temperature_rhs(
const std::pair<double, double> global_T_range,
const double global_max_velocity,
const double global_entropy_variation,
Assembly::Scratch::TemperatureRHS<dim> & scratch,
Assembly::CopyData::TemperatureRHS<dim> & data);
void copy_local_to_global_temperature_rhs(
const Assembly::CopyData::TemperatureRHS<dim> &data);
class Postprocessor;
};
template <int dim>
BoussinesqFlowProblem<dim>::Parameters::Parameters(
const std::string ¶meter_filename)
: end_time(1e8)
, initial_global_refinement(2)
, initial_adaptive_refinement(2)
, adaptive_refinement_interval(10)
, stabilization_alpha(2)
, stabilization_c_R(0.11)
, stabilization_beta(0.078)
, stokes_velocity_degree(2)
, use_locally_conservative_discretization(true)
, temperature_degree(2)
{
BoussinesqFlowProblem<dim>::Parameters::declare_parameters(prm);
std::ifstream parameter_file(parameter_filename);
if (!parameter_file)
{
parameter_file.close();
std::ofstream parameter_out(parameter_filename);
false,
"Input parameter file <" + parameter_filename +
"> not found. Creating a template file of the same name."));
}
parse_parameters(prm);
}
template <int dim>
void BoussinesqFlowProblem<dim>::Parameters::declare_parameters(
{
"1e8",
"The end time of the simulation in years.");
"2",
"The number of global refinement steps performed on "
"the initial coarse mesh, before the problem is first "
"solved there.");
"2",
"The number of adaptive refinement steps performed after "
"initial global refinement.");
"10",
"The number of time steps after which the mesh is to be "
"adapted based on computed error indicators.");
"false",
"Whether graphical output is to be generated or not. "
"You may not want to get graphical output if the number "
"of processors is large.");
"50",
"The number of time steps between each generation of "
"graphical output files.");
{
"2",
"The exponent in the entropy viscosity stabilization.");
"0.11",
"The c_R factor in the entropy viscosity "
"stabilization.");
"0.078",
"The beta factor in the artificial viscosity "
"stabilization. An appropriate value for 2d is 0.052 "
"and 0.078 for 3d.");
}
{
"Stokes velocity polynomial degree",
"2",
"The polynomial degree to use for the velocity variables "
"in the Stokes system.");
"Temperature polynomial degree",
"2",
"The polynomial degree to use for the temperature variable.");
"Use locally conservative discretization",
"true",
"Whether to use a Stokes discretization that is locally "
"conservative at the expense of a larger number of degrees "
"of freedom, or to go with a cheaper discretization "
"that does not locally conserve mass (although it is "
"globally conservative.");
}
}
template <int dim>
void BoussinesqFlowProblem<dim>::Parameters::parse_parameters(
{
initial_global_refinement = prm.
get_integer(
"Initial global refinement");
initial_adaptive_refinement =
adaptive_refinement_interval =
generate_graphical_output = prm.
get_bool(
"Generate graphical output");
graphical_output_interval =
prm.
get_integer(
"Time steps between graphical output");
{
}
{
stokes_velocity_degree =
temperature_degree = prm.
get_integer(
"Temperature polynomial degree");
use_locally_conservative_discretization =
prm.
get_bool(
"Use locally conservative discretization");
}
}
template <int dim>
BoussinesqFlowProblem<dim>::BoussinesqFlowProblem(Parameters ¶meters_)
: parameters(parameters_)
,
,
global_Omega_diameter(0.)
,
mapping(4)
,
stokes_fe(
FE_Q<dim>(parameters.stokes_velocity_degree),
dim,
(parameters.use_locally_conservative_discretization ?
FE_DGP<dim>(parameters.stokes_velocity_degree - 1)) :
FE_Q<dim>(parameters.stokes_velocity_degree - 1))),
1)
,
,
temperature_fe(parameters.temperature_degree)
,
time_step(0)
, old_time_step(0)
, timestep_number(0)
, rebuild_stokes_matrix(true)
, rebuild_stokes_preconditioner(true)
, rebuild_temperature_matrices(true)
, rebuild_temperature_preconditioner(true)
,
computing_timer(MPI_COMM_WORLD,
pcout,
{}
template <int dim>
double BoussinesqFlowProblem<dim>::get_maximal_velocity() const
{
parameters.stokes_velocity_degree);
const unsigned int n_q_points = quadrature_formula.size();
stokes_fe,
quadrature_formula,
std::vector<Tensor<1, dim>> velocity_values(n_q_points);
double max_local_velocity = 0;
for (const auto &cell : stokes_dof_handler.active_cell_iterators())
if (cell->is_locally_owned())
{
fe_values.reinit(cell);
fe_values[velocities].get_function_values(stokes_solution,
velocity_values);
for (unsigned int q = 0; q < n_q_points; ++q)
max_local_velocity =
}
}
template <int dim>
double BoussinesqFlowProblem<dim>::get_cfl_number() const
{
parameters.stokes_velocity_degree);
const unsigned int n_q_points = quadrature_formula.size();
stokes_fe,
quadrature_formula,
std::vector<Tensor<1, dim>> velocity_values(n_q_points);
double max_local_cfl = 0;
for (const auto &cell : stokes_dof_handler.active_cell_iterators())
if (cell->is_locally_owned())
{
fe_values.reinit(cell);
fe_values[velocities].get_function_values(stokes_solution,
velocity_values);
double max_local_velocity = 1
e-10;
for (unsigned int q = 0; q < n_q_points; ++q)
max_local_velocity =
max_local_cfl =
std::max(max_local_cfl, max_local_velocity / cell->diameter());
}
}
template <int dim>
double BoussinesqFlowProblem<dim>::get_entropy_variation(
const double average_temperature) const
{
if (parameters.stabilization_alpha != 2)
return 1.;
const QGauss<dim> quadrature_formula(parameters.temperature_degree + 1);
const unsigned int n_q_points = quadrature_formula.size();
quadrature_formula,
std::vector<double> old_temperature_values(n_q_points);
std::vector<double> old_old_temperature_values(n_q_points);
entropy_integrated = 0;
for (const auto &cell : temperature_dof_handler.active_cell_iterators())
if (cell->is_locally_owned())
{
fe_values.reinit(cell);
fe_values.get_function_values(old_temperature_solution,
old_temperature_values);
fe_values.get_function_values(old_old_temperature_solution,
old_old_temperature_values);
for (unsigned int q = 0; q < n_q_points; ++q)
{
(old_temperature_values[q] + old_old_temperature_values[q]) / 2;
const double entropy =
((
T - average_temperature) * (
T - average_temperature));
min_entropy =
std::min(min_entropy, entropy);
max_entropy =
std::max(max_entropy, entropy);
area += fe_values.JxW(q);
entropy_integrated += fe_values.JxW(q) * entropy;
}
}
const double local_sums[2] = {entropy_integrated, area},
local_maxima[2] = {-min_entropy, max_entropy};
double global_sums[2], global_maxima[2];
const double average_entropy = global_sums[0] / global_sums[1];
const double entropy_diff =
std::max(global_maxima[1] - average_entropy,
average_entropy - (-global_maxima[0]));
return entropy_diff;
}
template <int dim>
std::pair<double, double>
BoussinesqFlowProblem<dim>::get_extrapolated_temperature_range() const
{
parameters.temperature_degree);
const unsigned int n_q_points = quadrature_formula.size();
temperature_fe,
quadrature_formula,
std::vector<double> old_temperature_values(n_q_points);
std::vector<double> old_old_temperature_values(n_q_points);
if (timestep_number != 0)
{
for (const auto &cell : temperature_dof_handler.active_cell_iterators())
if (cell->is_locally_owned())
{
fe_values.reinit(cell);
fe_values.get_function_values(old_temperature_solution,
old_temperature_values);
fe_values.get_function_values(old_old_temperature_solution,
old_old_temperature_values);
for (unsigned int q = 0; q < n_q_points; ++q)
{
const double temperature =
(1. + time_step / old_time_step) *
old_temperature_values[q] -
time_step / old_time_step * old_old_temperature_values[q];
min_local_temperature =
std::min(min_local_temperature, temperature);
max_local_temperature =
std::max(max_local_temperature, temperature);
}
}
}
else
{
for (const auto &cell : temperature_dof_handler.active_cell_iterators())
if (cell->is_locally_owned())
{
fe_values.reinit(cell);
fe_values.get_function_values(old_temperature_solution,
old_temperature_values);
for (unsigned int q = 0; q < n_q_points; ++q)
{
const double temperature = old_temperature_values[q];
min_local_temperature =
std::min(min_local_temperature, temperature);
max_local_temperature =
std::max(max_local_temperature, temperature);
}
}
}
double local_extrema[2] = {-min_local_temperature, max_local_temperature};
double global_extrema[2];
return std::make_pair(-global_extrema[0], global_extrema[1]);
}
template <int dim>
double BoussinesqFlowProblem<dim>::compute_viscosity(
const std::vector<double> & old_temperature,
const std::vector<double> & old_old_temperature,
const std::vector<double> & old_temperature_laplacians,
const std::vector<double> & old_old_temperature_laplacians,
const double global_u_infty,
const double global_T_variation,
const double average_temperature,
const double global_entropy_variation,
const double cell_diameter) const
{
if (global_u_infty == 0)
return 5
e-3 * cell_diameter;
const unsigned int n_q_points = old_temperature.size();
double max_residual = 0;
double max_velocity = 0;
for (unsigned int q = 0; q < n_q_points; ++q)
{
(old_velocity_values[q] + old_old_velocity_values[q]) / 2;
(old_strain_rates[q] + old_old_strain_rates[q]) / 2;
const double T = (old_temperature[q] + old_old_temperature[q]) / 2;
const double dT_dt =
(old_temperature[q] - old_old_temperature[q]) / old_time_step;
const double u_grad_T =
u * (old_temperature_grads[q] + old_old_temperature_grads[q]) / 2;
const double kappa_Delta_T =
EquationData::kappa *
(old_temperature_laplacians[q] + old_old_temperature_laplacians[q]) /
2;
((EquationData::radiogenic_heating * EquationData::density(
T) +
2 * EquationData::eta * strain_rate * strain_rate) /
(EquationData::density(
T) * EquationData::specific_heat));
double residual =
std::abs(dT_dt + u_grad_T - kappa_Delta_T -
gamma);
if (parameters.stabilization_alpha == 2)
residual *=
std::abs(
T - average_temperature);
max_residual =
std::max(residual, max_residual);
}
const double max_viscosity =
(parameters.stabilization_beta * max_velocity * cell_diameter);
if (timestep_number == 0)
return max_viscosity;
else
{
double entropy_viscosity;
if (parameters.stabilization_alpha == 2)
entropy_viscosity =
(parameters.stabilization_c_R * cell_diameter * cell_diameter *
max_residual / global_entropy_variation);
else
entropy_viscosity =
(parameters.stabilization_c_R * cell_diameter *
global_Omega_diameter * max_velocity * max_residual /
(global_u_infty * global_T_variation));
return std::min(max_viscosity, entropy_viscosity);
}
}
template <int dim>
void BoussinesqFlowProblem<dim>::setup_stokes_matrix(
const std::vector<IndexSet> &stokes_partitioning,
const std::vector<IndexSet> &stokes_relevant_partitioning)
{
stokes_matrix.clear();
stokes_partitioning,
stokes_relevant_partitioning,
MPI_COMM_WORLD);
for (unsigned int c = 0; c < dim + 1; ++c)
for (
unsigned int d = 0;
d < dim + 1; ++
d)
if (!((c == dim) && (
d == dim)))
else
coupling,
sp,
stokes_constraints,
false,
MPI_COMM_WORLD));
sp.compress();
stokes_matrix.reinit(sp);
}
template <int dim>
void BoussinesqFlowProblem<dim>::setup_stokes_preconditioner(
const std::vector<IndexSet> &stokes_partitioning,
const std::vector<IndexSet> &stokes_relevant_partitioning)
{
Amg_preconditioner.reset();
Mp_preconditioner.reset();
stokes_preconditioner_matrix.clear();
stokes_partitioning,
stokes_relevant_partitioning,
MPI_COMM_WORLD);
for (unsigned int c = 0; c < dim + 1; ++c)
for (
unsigned int d = 0;
d < dim + 1; ++
d)
else
coupling,
sp,
stokes_constraints,
false,
MPI_COMM_WORLD));
sp.compress();
stokes_preconditioner_matrix.reinit(sp);
}
template <int dim>
void BoussinesqFlowProblem<dim>::setup_temperature_matrices(
const IndexSet &temperature_partitioner,
const IndexSet &temperature_relevant_partitioner)
{
T_preconditioner.reset();
temperature_mass_matrix.clear();
temperature_stiffness_matrix.clear();
temperature_matrix.clear();
temperature_partitioner,
temperature_relevant_partitioner,
MPI_COMM_WORLD);
sp,
temperature_constraints,
false,
MPI_COMM_WORLD));
sp.compress();
temperature_matrix.reinit(sp);
temperature_mass_matrix.reinit(sp);
temperature_stiffness_matrix.reinit(sp);
}
template <int dim>
void BoussinesqFlowProblem<dim>::setup_dofs()
{
stokes_dof_handler.distribute_dofs(stokes_fe);
std::vector<unsigned int> stokes_sub_blocks(dim + 1, 0);
stokes_sub_blocks[dim] = 1;
temperature_dof_handler.distribute_dofs(temperature_fe);
const std::vector<types::global_dof_index> stokes_dofs_per_block =
const unsigned int n_u = stokes_dofs_per_block[0],
n_p = stokes_dofs_per_block[1],
n_T = temperature_dof_handler.n_dofs();
std::locale s = pcout.get_stream().getloc();
pcout.get_stream().imbue(std::locale(""));
pcout <<
"Number of active cells: " <<
triangulation.n_global_active_cells()
<<
" (on " <<
triangulation.n_levels() <<
" levels)" << std::endl
<< "Number of degrees of freedom: " << n_u + n_p + n_T << " (" << n_u
<< '+' << n_p << '+' << n_T << ')' << std::endl
<< std::endl;
pcout.get_stream().imbue(s);
std::vector<IndexSet> stokes_partitioning, stokes_relevant_partitioning;
temperature_relevant_partitioning(n_T);
{
IndexSet stokes_index_set = stokes_dof_handler.locally_owned_dofs();
stokes_partitioning.push_back(stokes_index_set.
get_view(0, n_u));
stokes_partitioning.push_back(stokes_index_set.
get_view(n_u, n_u + n_p));
stokes_relevant_set);
stokes_relevant_partitioning.push_back(
stokes_relevant_partitioning.push_back(
stokes_relevant_set.
get_view(n_u, n_u + n_p));
temperature_partitioning = temperature_dof_handler.locally_owned_dofs();
temperature_dof_handler, temperature_relevant_partitioning);
}
{
stokes_constraints.clear();
stokes_constraints.reinit(stokes_relevant_set);
stokes_constraints);
stokes_dof_handler,
0,
stokes_constraints,
stokes_fe.component_mask(velocity_components));
std::set<types::boundary_id> no_normal_flux_boundaries;
no_normal_flux_boundaries.insert(1);
0,
no_normal_flux_boundaries,
stokes_constraints,
mapping);
stokes_constraints.close();
}
{
temperature_constraints.clear();
temperature_constraints.reinit(temperature_relevant_partitioning);
temperature_constraints);
temperature_dof_handler,
0,
EquationData::TemperatureInitialValues<dim>(),
temperature_constraints);
temperature_dof_handler,
1,
EquationData::TemperatureInitialValues<dim>(),
temperature_constraints);
temperature_constraints.close();
}
setup_stokes_matrix(stokes_partitioning, stokes_relevant_partitioning);
setup_stokes_preconditioner(stokes_partitioning,
stokes_relevant_partitioning);
setup_temperature_matrices(temperature_partitioning,
temperature_relevant_partitioning);
stokes_rhs.reinit(stokes_partitioning,
stokes_relevant_partitioning,
MPI_COMM_WORLD,
true);
stokes_solution.reinit(stokes_relevant_partitioning, MPI_COMM_WORLD);
old_stokes_solution.reinit(stokes_solution);
temperature_rhs.reinit(temperature_partitioning,
temperature_relevant_partitioning,
MPI_COMM_WORLD,
true);
temperature_solution.reinit(temperature_relevant_partitioning,
MPI_COMM_WORLD);
old_temperature_solution.reinit(temperature_solution);
old_old_temperature_solution.reinit(temperature_solution);
rebuild_stokes_matrix = true;
rebuild_stokes_preconditioner = true;
rebuild_temperature_matrices = true;
rebuild_temperature_preconditioner = true;
}
template <int dim>
void BoussinesqFlowProblem<dim>::local_assemble_stokes_preconditioner(
Assembly::Scratch::StokesPreconditioner<dim> & scratch,
Assembly::CopyData::StokesPreconditioner<dim> & data)
{
const unsigned int dofs_per_cell = stokes_fe.n_dofs_per_cell();
const unsigned int n_q_points =
scratch.stokes_fe_values.n_quadrature_points;
scratch.stokes_fe_values.reinit(cell);
cell->get_dof_indices(data.local_dof_indices);
data.local_matrix = 0;
for (unsigned int q = 0; q < n_q_points; ++q)
{
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
scratch.grad_phi_u[k] =
scratch.stokes_fe_values[velocities].gradient(k, q);
scratch.phi_p[k] = scratch.stokes_fe_values[pressure].value(k, q);
}
for (unsigned int i = 0; i < dofs_per_cell; ++i)
for (unsigned int j = 0; j < dofs_per_cell; ++j)
data.local_matrix(i, j) +=
(EquationData::eta *
(1. / EquationData::eta) * EquationData::pressure_scaling *
EquationData::pressure_scaling *
(scratch.phi_p[i] * scratch.phi_p[j])) *
scratch.stokes_fe_values.JxW(q);
}
}
template <int dim>
void BoussinesqFlowProblem<dim>::copy_local_to_global_stokes_preconditioner(
const Assembly::CopyData::StokesPreconditioner<dim> &data)
{
stokes_constraints.distribute_local_to_global(data.local_matrix,
data.local_dof_indices,
stokes_preconditioner_matrix);
}
template <int dim>
void BoussinesqFlowProblem<dim>::assemble_stokes_preconditioner()
{
stokes_preconditioner_matrix = 0;
const QGauss<dim> quadrature_formula(parameters.stokes_velocity_degree + 1);
using CellFilter =
auto worker =
Assembly::Scratch::StokesPreconditioner<dim> & scratch,
Assembly::CopyData::StokesPreconditioner<dim> & data) {
this->local_assemble_stokes_preconditioner(cell, scratch, data);
};
auto copier =
[this](const Assembly::CopyData::StokesPreconditioner<dim> &data) {
this->copy_local_to_global_stokes_preconditioner(data);
};
stokes_dof_handler.begin_active()),
stokes_dof_handler.end()),
worker,
copier,
Assembly::Scratch::StokesPreconditioner<dim>(
stokes_fe,
quadrature_formula,
mapping,
Assembly::CopyData::StokesPreconditioner<dim>(stokes_fe));
}
template <int dim>
void BoussinesqFlowProblem<dim>::build_stokes_preconditioner()
{
if (rebuild_stokes_preconditioner == false)
return;
" Build Stokes preconditioner");
pcout << " Rebuilding Stokes preconditioner..." << std::flush;
assemble_stokes_preconditioner();
std::vector<std::vector<bool>> constant_modes;
stokes_fe.component_mask(
velocity_components),
constant_modes);
Mp_preconditioner =
std::make_shared<TrilinosWrappers::PreconditionJacobi>();
Amg_preconditioner = std::make_shared<TrilinosWrappers::PreconditionAMG>();
Mp_preconditioner->initialize(stokes_preconditioner_matrix.block(1, 1));
Amg_preconditioner->initialize(stokes_preconditioner_matrix.block(0, 0),
Amg_data);
rebuild_stokes_preconditioner = false;
pcout << std::endl;
}
template <int dim>
void BoussinesqFlowProblem<dim>::local_assemble_stokes_system(
Assembly::Scratch::StokesSystem<dim> & scratch,
Assembly::CopyData::StokesSystem<dim> & data)
{
const unsigned int dofs_per_cell =
scratch.stokes_fe_values.get_fe().n_dofs_per_cell();
const unsigned int n_q_points =
scratch.stokes_fe_values.n_quadrature_points;
scratch.stokes_fe_values.reinit(cell);
&
triangulation, cell->level(), cell->index(), &temperature_dof_handler);
scratch.temperature_fe_values.reinit(temperature_cell);
if (rebuild_stokes_matrix)
data.local_matrix = 0;
data.local_rhs = 0;
scratch.temperature_fe_values.get_function_values(
old_temperature_solution, scratch.old_temperature_values);
for (unsigned int q = 0; q < n_q_points; ++q)
{
const double old_temperature = scratch.old_temperature_values[q];
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
scratch.phi_u[k] = scratch.stokes_fe_values[velocities].value(k, q);
if (rebuild_stokes_matrix)
{
scratch.grads_phi_u[k] =
scratch.stokes_fe_values[velocities].symmetric_gradient(k, q);
scratch.div_phi_u[k] =
scratch.stokes_fe_values[velocities].divergence(k, q);
scratch.phi_p[k] =
scratch.stokes_fe_values[pressure].value(k, q);
}
}
if (rebuild_stokes_matrix == true)
for (unsigned int i = 0; i < dofs_per_cell; ++i)
for (unsigned int j = 0; j < dofs_per_cell; ++j)
data.local_matrix(i, j) +=
(EquationData::eta * 2 *
(scratch.grads_phi_u[i] * scratch.grads_phi_u[j]) -
(EquationData::pressure_scaling * scratch.div_phi_u[i] *
scratch.phi_p[j]) -
(EquationData::pressure_scaling * scratch.phi_p[i] *
scratch.div_phi_u[j])) *
scratch.stokes_fe_values.JxW(q);
scratch.stokes_fe_values.quadrature_point(q));
for (unsigned int i = 0; i < dofs_per_cell; ++i)
data.local_rhs(i) += (EquationData::density(old_temperature) *
gravity * scratch.phi_u[i]) *
scratch.stokes_fe_values.JxW(q);
}
cell->get_dof_indices(data.local_dof_indices);
}
template <int dim>
void BoussinesqFlowProblem<dim>::copy_local_to_global_stokes_system(
const Assembly::CopyData::StokesSystem<dim> &data)
{
if (rebuild_stokes_matrix == true)
stokes_constraints.distribute_local_to_global(data.local_matrix,
data.local_rhs,
data.local_dof_indices,
stokes_matrix,
stokes_rhs);
else
stokes_constraints.distribute_local_to_global(data.local_rhs,
data.local_dof_indices,
stokes_rhs);
}
template <int dim>
void BoussinesqFlowProblem<dim>::assemble_stokes_system()
{
" Assemble Stokes system");
if (rebuild_stokes_matrix == true)
stokes_matrix = 0;
stokes_rhs = 0;
const QGauss<dim> quadrature_formula(parameters.stokes_velocity_degree + 1);
using CellFilter =
stokes_dof_handler.begin_active()),
Assembly::Scratch::StokesSystem<dim> & scratch,
Assembly::CopyData::StokesSystem<dim> & data) {
this->local_assemble_stokes_system(cell, scratch, data);
},
[this](const Assembly::CopyData::StokesSystem<dim> &data) {
this->copy_local_to_global_stokes_system(data);
},
Assembly::Scratch::StokesSystem<dim>(
stokes_fe,
mapping,
quadrature_formula,
temperature_fe,
Assembly::CopyData::StokesSystem<dim>(stokes_fe));
if (rebuild_stokes_matrix == true)
rebuild_stokes_matrix = false;
pcout << std::endl;
}
template <int dim>
void BoussinesqFlowProblem<dim>::local_assemble_temperature_matrix(
Assembly::Scratch::TemperatureMatrix<dim> & scratch,
Assembly::CopyData::TemperatureMatrix<dim> & data)
{
const unsigned int dofs_per_cell =
scratch.temperature_fe_values.get_fe().n_dofs_per_cell();
const unsigned int n_q_points =
scratch.temperature_fe_values.n_quadrature_points;
scratch.temperature_fe_values.reinit(cell);
cell->get_dof_indices(data.local_dof_indices);
data.local_mass_matrix = 0;
data.local_stiffness_matrix = 0;
for (unsigned int q = 0; q < n_q_points; ++q)
{
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
scratch.grad_phi_T[k] =
scratch.temperature_fe_values.shape_grad(k, q);
scratch.phi_T[k] = scratch.temperature_fe_values.shape_value(k, q);
}
for (unsigned int i = 0; i < dofs_per_cell; ++i)
for (unsigned int j = 0; j < dofs_per_cell; ++j)
{
data.local_mass_matrix(i, j) +=
(scratch.phi_T[i] * scratch.phi_T[j] *
scratch.temperature_fe_values.JxW(q));
data.local_stiffness_matrix(i, j) +=
(EquationData::kappa * scratch.grad_phi_T[i] *
scratch.grad_phi_T[j] * scratch.temperature_fe_values.JxW(q));
}
}
}
template <int dim>
void BoussinesqFlowProblem<dim>::copy_local_to_global_temperature_matrix(
const Assembly::CopyData::TemperatureMatrix<dim> &data)
{
temperature_constraints.distribute_local_to_global(data.local_mass_matrix,
data.local_dof_indices,
temperature_mass_matrix);
temperature_constraints.distribute_local_to_global(
data.local_stiffness_matrix,
data.local_dof_indices,
temperature_stiffness_matrix);
}
template <int dim>
void BoussinesqFlowProblem<dim>::assemble_temperature_matrix()
{
if (rebuild_temperature_matrices == false)
return;
" Assemble temperature matrices");
temperature_mass_matrix = 0;
temperature_stiffness_matrix = 0;
const QGauss<dim> quadrature_formula(parameters.temperature_degree + 2);
using CellFilter =
temperature_dof_handler.begin_active()),
temperature_dof_handler.end()),
Assembly::Scratch::TemperatureMatrix<dim> & scratch,
Assembly::CopyData::TemperatureMatrix<dim> & data) {
this->local_assemble_temperature_matrix(cell, scratch, data);
},
[this](const Assembly::CopyData::TemperatureMatrix<dim> &data) {
this->copy_local_to_global_temperature_matrix(data);
},
Assembly::Scratch::TemperatureMatrix<dim>(temperature_fe,
mapping,
quadrature_formula),
Assembly::CopyData::TemperatureMatrix<dim>(temperature_fe));
rebuild_temperature_matrices = false;
rebuild_temperature_preconditioner = true;
}
template <int dim>
void BoussinesqFlowProblem<dim>::local_assemble_temperature_rhs(
const std::pair<double, double> global_T_range,
const double global_max_velocity,
const double global_entropy_variation,
Assembly::Scratch::TemperatureRHS<dim> & scratch,
Assembly::CopyData::TemperatureRHS<dim> & data)
{
const bool use_bdf2_scheme = (timestep_number != 0);
const unsigned int dofs_per_cell =
scratch.temperature_fe_values.get_fe().n_dofs_per_cell();
const unsigned int n_q_points =
scratch.temperature_fe_values.n_quadrature_points;
data.local_rhs = 0;
data.matrix_for_bc = 0;
cell->get_dof_indices(data.local_dof_indices);
scratch.temperature_fe_values.
reinit(cell);
&
triangulation, cell->level(), cell->index(), &stokes_dof_handler);
scratch.stokes_fe_values.reinit(stokes_cell);
scratch.temperature_fe_values.get_function_values(
old_temperature_solution, scratch.old_temperature_values);
scratch.temperature_fe_values.get_function_values(
old_old_temperature_solution, scratch.old_old_temperature_values);
scratch.temperature_fe_values.get_function_gradients(
old_temperature_solution, scratch.old_temperature_grads);
scratch.temperature_fe_values.get_function_gradients(
old_old_temperature_solution, scratch.old_old_temperature_grads);
scratch.temperature_fe_values.get_function_laplacians(
old_temperature_solution, scratch.old_temperature_laplacians);
scratch.temperature_fe_values.get_function_laplacians(
old_old_temperature_solution, scratch.old_old_temperature_laplacians);
scratch.stokes_fe_values[velocities].get_function_values(
stokes_solution, scratch.old_velocity_values);
scratch.stokes_fe_values[velocities].get_function_values(
old_stokes_solution, scratch.old_old_velocity_values);
scratch.stokes_fe_values[velocities].get_function_symmetric_gradients(
stokes_solution, scratch.old_strain_rates);
scratch.stokes_fe_values[velocities].get_function_symmetric_gradients(
old_stokes_solution, scratch.old_old_strain_rates);
const double nu =
compute_viscosity(scratch.old_temperature_values,
scratch.old_old_temperature_values,
scratch.old_temperature_grads,
scratch.old_old_temperature_grads,
scratch.old_temperature_laplacians,
scratch.old_old_temperature_laplacians,
scratch.old_velocity_values,
scratch.old_old_velocity_values,
scratch.old_strain_rates,
scratch.old_old_strain_rates,
global_max_velocity,
global_T_range.second - global_T_range.first,
0.5 * (global_T_range.second + global_T_range.first),
global_entropy_variation,
cell->diameter());
for (unsigned int q = 0; q < n_q_points; ++q)
{
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
scratch.phi_T[k] = scratch.temperature_fe_values.shape_value(k, q);
scratch.grad_phi_T[k] =
scratch.temperature_fe_values.shape_grad(k, q);
}
const double T_term_for_rhs =
(use_bdf2_scheme ?
(scratch.old_temperature_values[q] *
(1 + time_step / old_time_step) -
scratch.old_old_temperature_values[q] * (time_step * time_step) /
(old_time_step * (time_step + old_time_step))) :
scratch.old_temperature_values[q]);
const double ext_T =
(use_bdf2_scheme ? (scratch.old_temperature_values[q] *
(1 + time_step / old_time_step) -
scratch.old_old_temperature_values[q] *
time_step / old_time_step) :
scratch.old_temperature_values[q]);
(use_bdf2_scheme ? (scratch.old_temperature_grads[q] *
(1 + time_step / old_time_step) -
scratch.old_old_temperature_grads[q] * time_step /
old_time_step) :
scratch.old_temperature_grads[q]);
(use_bdf2_scheme ?
(scratch.old_velocity_values[q] * (1 + time_step / old_time_step) -
scratch.old_old_velocity_values[q] * time_step / old_time_step) :
scratch.old_velocity_values[q]);
(use_bdf2_scheme ?
(scratch.old_strain_rates[q] * (1 + time_step / old_time_step) -
scratch.old_old_strain_rates[q] * time_step / old_time_step) :
scratch.old_strain_rates[q]);
((EquationData::radiogenic_heating * EquationData::density(ext_T) +
2 * EquationData::eta * extrapolated_strain_rate *
extrapolated_strain_rate) /
(EquationData::density(ext_T) * EquationData::specific_heat));
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
data.local_rhs(i) +=
(T_term_for_rhs * scratch.phi_T[i] -
time_step * extrapolated_u * ext_grad_T * scratch.phi_T[i] -
time_step * nu * ext_grad_T * scratch.grad_phi_T[i] +
time_step *
gamma * scratch.phi_T[i]) *
scratch.temperature_fe_values.JxW(q);
if (temperature_constraints.is_inhomogeneously_constrained(
data.local_dof_indices[i]))
{
for (unsigned int j = 0; j < dofs_per_cell; ++j)
data.matrix_for_bc(j, i) +=
(scratch.phi_T[i] * scratch.phi_T[j] *
(use_bdf2_scheme ? ((2 * time_step + old_time_step) /
(time_step + old_time_step)) :
1.) +
scratch.grad_phi_T[i] * scratch.grad_phi_T[j] *
EquationData::kappa * time_step) *
scratch.temperature_fe_values.JxW(q);
}
}
}
}
template <int dim>
void BoussinesqFlowProblem<dim>::copy_local_to_global_temperature_rhs(
const Assembly::CopyData::TemperatureRHS<dim> &data)
{
temperature_constraints.distribute_local_to_global(data.local_rhs,
data.local_dof_indices,
temperature_rhs,
data.matrix_for_bc);
}
template <int dim>
void BoussinesqFlowProblem<dim>::assemble_temperature_system(
const double maximal_velocity)
{
const bool use_bdf2_scheme = (timestep_number != 0);
if (use_bdf2_scheme == true)
{
temperature_matrix.copy_from(temperature_mass_matrix);
temperature_matrix *=
(2 * time_step + old_time_step) / (time_step + old_time_step);
temperature_matrix.add(time_step, temperature_stiffness_matrix);
}
else
{
temperature_matrix.copy_from(temperature_mass_matrix);
temperature_matrix.add(time_step, temperature_stiffness_matrix);
}
if (rebuild_temperature_preconditioner == true)
{
T_preconditioner =
std::make_shared<TrilinosWrappers::PreconditionJacobi>();
T_preconditioner->initialize(temperature_matrix);
rebuild_temperature_preconditioner = false;
}
temperature_rhs = 0;
const QGauss<dim> quadrature_formula(parameters.temperature_degree + 2);
const std::pair<double, double> global_T_range =
get_extrapolated_temperature_range();
const double average_temperature =
0.5 * (global_T_range.first + global_T_range.second);
const double global_entropy_variation =
get_entropy_variation(average_temperature);
using CellFilter =
auto worker =
[this, global_T_range, maximal_velocity, global_entropy_variation](
Assembly::Scratch::TemperatureRHS<dim> & scratch,
Assembly::CopyData::TemperatureRHS<dim> & data) {
this->local_assemble_temperature_rhs(global_T_range,
maximal_velocity,
global_entropy_variation,
cell,
scratch,
data);
};
auto copier = [this](const Assembly::CopyData::TemperatureRHS<dim> &data) {
this->copy_local_to_global_temperature_rhs(data);
};
temperature_dof_handler.begin_active()),
temperature_dof_handler.end()),
worker,
copier,
Assembly::Scratch::TemperatureRHS<dim>(
temperature_fe, stokes_fe, mapping, quadrature_formula),
Assembly::CopyData::TemperatureRHS<dim>(temperature_fe));
}
template <int dim>
void BoussinesqFlowProblem<dim>::solve()
{
{
" Solve Stokes system");
pcout << " Solving Stokes system... " << std::flush;
stokes_rhs);
distributed_stokes_solution = stokes_solution;
distributed_stokes_solution.block(1) /= EquationData::pressure_scaling;
const unsigned int
start = (distributed_stokes_solution.block(0).size() +
distributed_stokes_solution.block(1).local_range().first),
end = (distributed_stokes_solution.block(0).size() +
distributed_stokes_solution.block(1).local_range().second);
for (
unsigned int i = start; i <
end; ++i)
if (stokes_constraints.is_constrained(i))
distributed_stokes_solution(i) = 0;
unsigned int n_iterations = 0;
const double solver_tolerance = 1
e-8 * stokes_rhs.l2_norm();
try
{
const LinearSolvers::BlockSchurPreconditioner<
preconditioner(stokes_matrix,
stokes_preconditioner_matrix,
*Mp_preconditioner,
*Amg_preconditioner,
false);
solver_control,
mem,
30));
solver.solve(stokes_matrix,
distributed_stokes_solution,
stokes_rhs,
preconditioner);
n_iterations = solver_control.last_step();
}
{
const LinearSolvers::BlockSchurPreconditioner<
preconditioner(stokes_matrix,
stokes_preconditioner_matrix,
*Mp_preconditioner,
*Amg_preconditioner,
true);
solver_tolerance);
solver_control_refined,
mem,
50));
solver.solve(stokes_matrix,
distributed_stokes_solution,
stokes_rhs,
preconditioner);
n_iterations =
(solver_control.last_step() + solver_control_refined.last_step());
}
stokes_constraints.distribute(distributed_stokes_solution);
distributed_stokes_solution.block(1) *= EquationData::pressure_scaling;
stokes_solution = distributed_stokes_solution;
pcout << n_iterations << " iterations." << std::endl;
}
{
" Assemble temperature rhs");
old_time_step = time_step;
const double scaling = (dim == 3 ? 0.25 : 1.0);
time_step = (scaling / (2.1 * dim *
std::sqrt(1. * dim)) /
(parameters.temperature_degree * get_cfl_number()));
const double maximal_velocity = get_maximal_velocity();
pcout << " Maximal velocity: "
<< maximal_velocity * EquationData::year_in_seconds * 100
<< " cm/year" << std::endl;
pcout << " "
<< "Time step: " << time_step / EquationData::year_in_seconds
<< " years" << std::endl;
temperature_solution = old_temperature_solution;
assemble_temperature_system(maximal_velocity);
}
{
" Solve temperature system");
1
e-12 * temperature_rhs.l2_norm());
temperature_rhs);
distributed_temperature_solution = temperature_solution;
cg.solve(temperature_matrix,
distributed_temperature_solution,
temperature_rhs,
*T_preconditioner);
temperature_constraints.distribute(distributed_temperature_solution);
temperature_solution = distributed_temperature_solution;
pcout << " " << solver_control.last_step()
<< " CG iterations for temperature" << std::endl;
double global_temperature[2];
for (unsigned int i =
distributed_temperature_solution.local_range().first;
i < distributed_temperature_solution.local_range().second;
++i)
{
temperature[0] =
std::min<double>(temperature[0],
distributed_temperature_solution(i));
temperature[1] =
std::max<double>(temperature[1],
distributed_temperature_solution(i));
}
temperature[0] *= -1.0;
global_temperature[0] *= -1.0;
pcout << " Temperature range: " << global_temperature[0] << ' '
<< global_temperature[1] << std::endl;
}
}
template <int dim>
class BoussinesqFlowProblem<dim>::Postprocessor
{
public:
Postprocessor(
const unsigned int partition,
const double minimal_pressure);
virtual void evaluate_vector_field(
virtual std::vector<std::string> get_names() const override;
virtual std::vector<
get_data_component_interpretation() const override;
virtual UpdateFlags get_needed_update_flags()
const override;
private:
const double minimal_pressure;
};
template <int dim>
BoussinesqFlowProblem<dim>::Postprocessor::Postprocessor(
const double minimal_pressure)
, minimal_pressure(minimal_pressure)
{}
template <int dim>
std::vector<std::string>
BoussinesqFlowProblem<dim>::Postprocessor::get_names() const
{
std::vector<std::string> solution_names(dim, "velocity");
solution_names.emplace_back("p");
solution_names.emplace_back("T");
solution_names.emplace_back("friction_heating");
solution_names.emplace_back("partition");
return solution_names;
}
template <int dim>
std::vector<DataComponentInterpretation::DataComponentInterpretation>
BoussinesqFlowProblem<dim>::Postprocessor::get_data_component_interpretation()
const
{
std::vector<DataComponentInterpretation::DataComponentInterpretation>
interpretation(dim,
return interpretation;
}
template <int dim>
BoussinesqFlowProblem<dim>::Postprocessor::get_needed_update_flags() const
{
}
template <int dim>
void BoussinesqFlowProblem<dim>::Postprocessor::evaluate_vector_field(
{
Assert(computed_quantities.size() == n_quadrature_points,
for (unsigned int q = 0; q < n_quadrature_points; ++q)
{
for (
unsigned int d = 0;
d < dim; ++
d)
EquationData::year_in_seconds * 100);
const double pressure =
computed_quantities[q](dim) = pressure;
computed_quantities[q](dim + 1) = temperature;
for (
unsigned int d = 0;
d < dim; ++
d)
computed_quantities[q](dim + 2) =
2 * EquationData::eta * strain_rate * strain_rate;
}
}
template <int dim>
void BoussinesqFlowProblem<dim>::output_results()
{
joint_dof_handler.distribute_dofs(joint_fe);
Assert(joint_dof_handler.n_dofs() ==
stokes_dof_handler.n_dofs() + temperature_dof_handler.n_dofs(),
joint_solution.
reinit(joint_dof_handler.locally_owned_dofs(),
MPI_COMM_WORLD);
{
std::vector<types::global_dof_index> local_joint_dof_indices(
joint_fe.n_dofs_per_cell());
std::vector<types::global_dof_index> local_stokes_dof_indices(
stokes_fe.n_dofs_per_cell());
std::vector<types::global_dof_index> local_temperature_dof_indices(
temperature_fe.n_dofs_per_cell());
joint_cell = joint_dof_handler.begin_active(),
joint_endc = joint_dof_handler.end(),
stokes_cell = stokes_dof_handler.begin_active(),
temperature_cell = temperature_dof_handler.begin_active();
for (; joint_cell != joint_endc;
++joint_cell, ++stokes_cell, ++temperature_cell)
if (joint_cell->is_locally_owned())
{
joint_cell->get_dof_indices(local_joint_dof_indices);
stokes_cell->get_dof_indices(local_stokes_dof_indices);
temperature_cell->get_dof_indices(local_temperature_dof_indices);
for (unsigned int i = 0; i < joint_fe.n_dofs_per_cell(); ++i)
if (joint_fe.system_to_base_index(i).first.first == 0)
{
Assert(joint_fe.system_to_base_index(i).second <
local_stokes_dof_indices.size(),
joint_solution(local_joint_dof_indices[i]) = stokes_solution(
local_stokes_dof_indices[joint_fe.system_to_base_index(i)
.second]);
}
else
{
Assert(joint_fe.system_to_base_index(i).first.first == 1,
Assert(joint_fe.system_to_base_index(i).second <
local_temperature_dof_indices.size(),
joint_solution(local_joint_dof_indices[i]) =
temperature_solution(
local_temperature_dof_indices
[joint_fe.system_to_base_index(i).second]);
}
}
}
IndexSet locally_relevant_joint_dofs(joint_dof_handler.n_dofs());
locally_relevant_joint_dofs);
locally_relevant_joint_solution.
reinit(locally_relevant_joint_dofs,
MPI_COMM_WORLD);
locally_relevant_joint_solution = joint_solution;
MPI_COMM_WORLD),
stokes_solution.block(1).min());
static int out_index = 0;
"./", "solution", out_index, MPI_COMM_WORLD, 5);
out_index++;
}
template <int dim>
void
BoussinesqFlowProblem<dim>::refine_mesh(const unsigned int max_grid_level)
{
temperature_trans(temperature_dof_handler);
stokes_trans(stokes_dof_handler);
{
"Refine mesh structure, part 1");
temperature_dof_handler,
temperature_solution,
estimated_error_per_cell,
nullptr,
0,
++cell)
cell->clear_refine_flag();
std::vector<const TrilinosWrappers::MPI::Vector *> x_temperature(2);
x_temperature[0] = &temperature_solution;
x_temperature[1] = &old_temperature_solution;
std::vector<const TrilinosWrappers::MPI::BlockVector *> x_stokes(2);
x_stokes[0] = &stokes_solution;
x_stokes[1] = &old_stokes_solution;
temperature_trans.prepare_for_coarsening_and_refinement(x_temperature);
stokes_trans.prepare_for_coarsening_and_refinement(x_stokes);
}
setup_dofs();
{
"Refine mesh structure, part 2");
{
std::vector<TrilinosWrappers::MPI::Vector *> tmp(2);
tmp[0] = &(distributed_temp1);
tmp[1] = &(distributed_temp2);
temperature_trans.interpolate(tmp);
temperature_constraints.distribute(distributed_temp1);
temperature_constraints.distribute(distributed_temp2);
temperature_solution = distributed_temp1;
old_temperature_solution = distributed_temp2;
}
{
std::vector<TrilinosWrappers::MPI::BlockVector *> stokes_tmp(2);
stokes_tmp[0] = &(distributed_stokes);
stokes_tmp[1] = &(old_distributed_stokes);
stokes_trans.interpolate(stokes_tmp);
stokes_constraints.distribute(distributed_stokes);
stokes_constraints.distribute(old_distributed_stokes);
stokes_solution = distributed_stokes;
old_stokes_solution = old_distributed_stokes;
}
}
}
template <int dim>
{
EquationData::R0,
EquationData::R1,
(dim == 3) ? 96 : 12,
true);
triangulation.refine_global(parameters.initial_global_refinement);
setup_dofs();
unsigned int pre_refinement_step = 0;
start_time_iteration:
{
temperature_dof_handler.locally_owned_dofs());
temperature_constraints,
EquationData::TemperatureInitialValues<dim>(),
solution);
temperature_solution = solution;
old_temperature_solution = solution;
old_old_temperature_solution = solution;
}
timestep_number = 0;
time_step = old_time_step = 0;
double time = 0;
do
{
pcout << "Timestep " << timestep_number
<< ": t=" << time / EquationData::year_in_seconds << " years"
<< std::endl;
assemble_stokes_system();
build_stokes_preconditioner();
assemble_temperature_matrix();
solve();
pcout << std::endl;
if ((timestep_number == 0) &&
(pre_refinement_step < parameters.initial_adaptive_refinement))
{
refine_mesh(parameters.initial_global_refinement +
parameters.initial_adaptive_refinement);
++pre_refinement_step;
goto start_time_iteration;
}
else if ((timestep_number > 0) &&
(timestep_number % parameters.adaptive_refinement_interval ==
0))
refine_mesh(parameters.initial_global_refinement +
parameters.initial_adaptive_refinement);
if ((parameters.generate_graphical_output == true) &&
(timestep_number % parameters.graphical_output_interval == 0))
output_results();
if (time > parameters.end_time * EquationData::year_in_seconds)
break;
old_old_stokes_solution = old_stokes_solution;
old_stokes_solution = stokes_solution;
old_old_temperature_solution = old_temperature_solution;
old_temperature_solution = temperature_solution;
if (old_time_step > 0)
{
{
distr_solution = stokes_solution;
distr_old_solution = old_old_stokes_solution;
distr_solution.
sadd(1. + time_step / old_time_step,
-time_step / old_time_step,
distr_old_solution);
stokes_solution = distr_solution;
}
{
distr_solution = temperature_solution;
distr_old_solution = old_old_temperature_solution;
distr_solution.sadd(1. + time_step / old_time_step,
-time_step / old_time_step,
distr_old_solution);
temperature_solution = distr_solution;
}
}
if ((timestep_number > 0) && (timestep_number % 100 == 0))
computing_timer.print_summary();
time += time_step;
++timestep_number;
}
while (true);
if ((parameters.generate_graphical_output == true) &&
!((timestep_number - 1) % parameters.graphical_output_interval == 0))
output_results();
}
}
int main(int argc, char *argv[])
{
try
{
using namespace Step32;
std::string parameter_filename;
if (argc >= 2)
parameter_filename = argv[1];
else
parameter_filename = "step-32.prm";
const int dim = 2;
BoussinesqFlowProblem<dim>::Parameters parameters(parameter_filename);
BoussinesqFlowProblem<dim> flow_problem(parameters);
flow_problem.run();
}
catch (std::exception &exc)
{
std::cerr << std::endl
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Exception on processing: " << std::endl
<< exc.what() << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
catch (...)
{
std::cerr << std::endl
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Unknown exception!" << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
return 0;
}
::VectorizedArray< Number, width > cos(const ::VectorizedArray< Number, width > &)