This program was contributed by Jean-Paul Pelteret <jppelteret@gmail.com>.
This program is part of the deal.II code gallery and consists of the following files (click to inspect):
Pictures from this code gallery program
Annotated version of README.md
Overview
The complex interaction of muscles with their surrounding anatomy and environment plays a vital role in the many activities that are required for animals to live and survive. Skeletal muscle composes a large portion of that musculo-skeletal system, and is controlled by the central nervous system in a conscious or unconscious manner. For humans in particular, the construction of- and control mechanisms behind skeletal muscle allows us to accomplish complex tasks ranging from those that are physically exerting to those that are delicate and require great dexterity.
As an introduction into the biomechanics of the human muscular system, we combine a well known load-activation pattern taken from well established literature on the topic (both in the fields of human physiology and the computational simulation thereof) with an idealised model of a part of the human anatomy that most can easily identify with, namely the biceps brachii.
An idealised model of the human biceps brachii
To tackle this problem, we do not deviate particularly far from the approach that is comprehensively documented in @ref step_8 "step-8". The primary differences between this code-gallery example and the tutorial is the alteration of the geometry and boundary conditions that accompany it, as well as the extension of the constitutive law to include the transversely isotropic active muscle model. We thus present both the theory and associated nomenclature (which is mimicked in the code itself) in the accompanying document. There you can observe the additional contributions to both the left- and right-hand sides of the linear system due to the integration of the rate-dependent, linearised muscle model. Although such linear model models are valid only under conditions of small deformations, in this scenario we will (with a healthy dose of skepticism) make a very coarse-grained first approximation of the muscles behaviour in the large deformation regime.
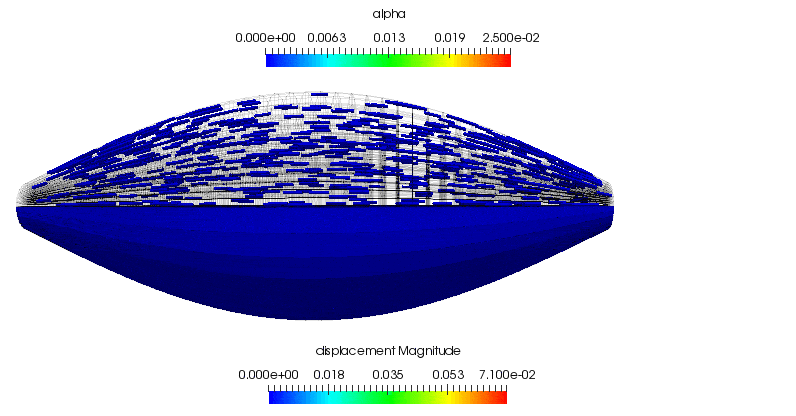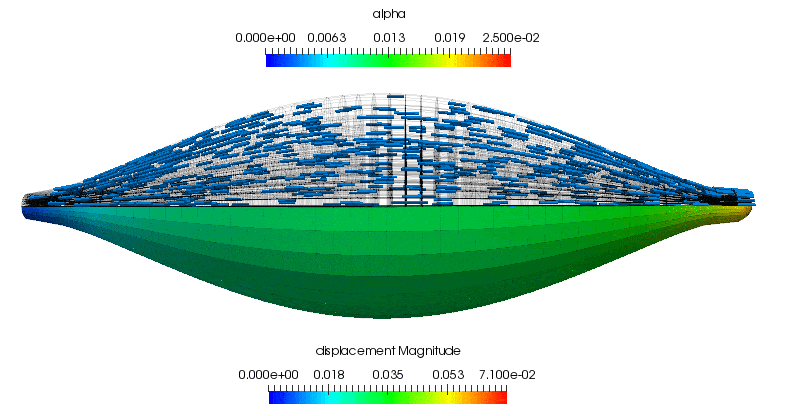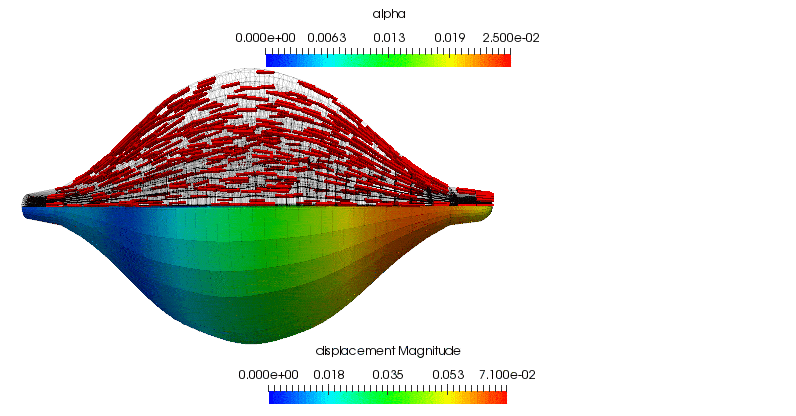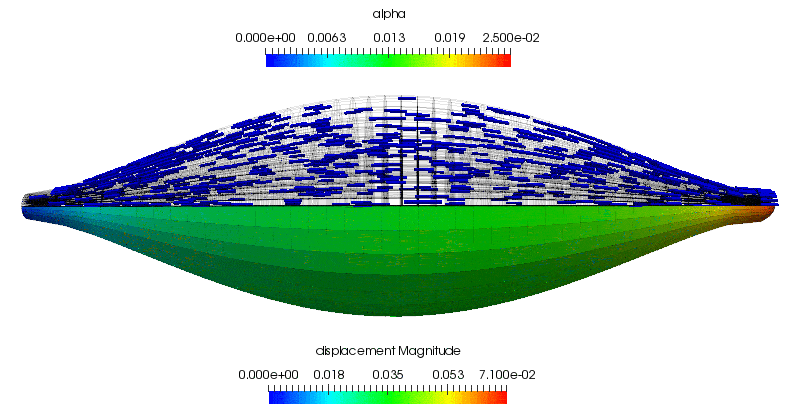
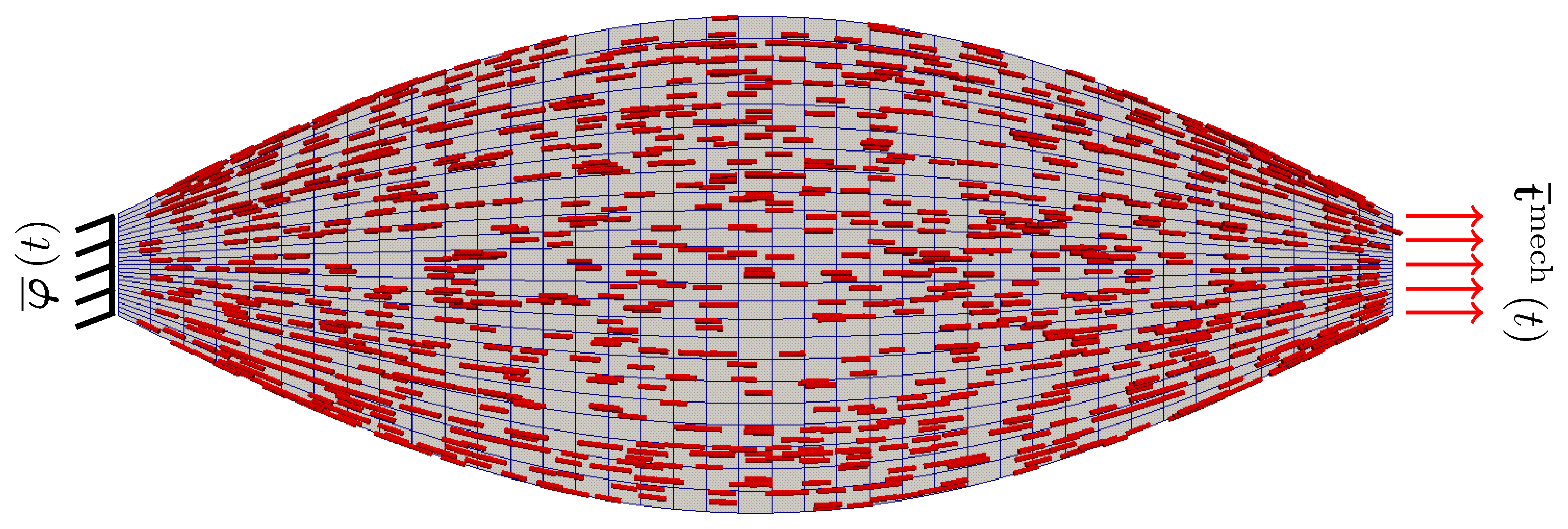
The basic problem configuration, including a depiction of the underlying muscle microstructural orientation, is (loosely) summarised in the following image.
Note that the driver for the deformation of the muscle tissue are the applied traction alone when the muscle is in a passive state. However, during active contraction, as governed by the prescribed input neural signal, the muscle works against the applied traction. This condition, where the traction applied to a muscle is constant during periods of muscle activation, is known as isotonic contraction. More specifically, since overall the muscle shortens during contraction we are in fact modelling concentric contraction of the biceps.
As for the specific geometry of the problem, we consider an idealised human biceps with a length of 250mm, insertion and origin diameter of 20mm and a diameter of 80mm at its mid-point. We assume that there exists a single muscle fibre family orientated axially. The orientation of the underlying muscle fibres is, however, not parallel, but rather follows the curvature of the macroscopic anatomy. The longitudinal profile of the muscle is generated using a trignometric function, as opposed to being extracted from medical images. The benefit to doing so is that the geometry can be (parametrically) created in deal.II itself and the associated microstructural orientation can be directly linked to the user-defined geometry.
Requirements
Version 8.5 or greater of deal.II
There are no other requirements with regards to the third-party packages that deal.II can link to.
Compiling and running
Similar to the example programs, run
cmake -DDEAL_II_DIR=/path/to/deal.II .
in this directory to configure the problem.
or
The problem may then be run with
Some simulation parameters may be changed by adjusting the parameters.prm file. Notably, its possible to switch between the model of the biceps and the reduced geometry used to reproduce the linearised counterpart of the isotonic contraction numerical experiments conducted by Martins.
Reference for this work
If you use this program as a basis for your own work, please consider citing it in your list of references. The initial version of this work was contributed to the deal.II project by the authors listed in the following citation: J-P. V. Pelteret and T. Hamann, The deal.II code gallery: Linear Elastic Active Skeletal Muscle Mode, 2017. DOI: 10.5281/zenodo.437608
Acknowledgements
The support of this work by the European Research Council (ERC) through the Advanced Grant 289049 MOCOPOLY is gratefully acknowledged by the first author.
Recommended Literature
Kajee, Y. and Pelteret, J-P. V. and Reddy, B. D. (2013), The biomechanics of the human tongue. International Journal for Numerical Methods in Biomedical Engineering 29 , 4, 492-514. DOI: 10.1002/cnm.2531 ;
J-P. V. Pelteret, A computational neuromuscular model of the human upper airway with application to the study of obstructive sleep apnoea. PhD Thesis, University of Cape Town, 2013. http://hdl.handle.net/11427/9519 ;
Martins, J. A. C. and Pires, E. B. and Salvado, R. and Dinis, P. B. (1998), A numerical model of passive and active behaviour of skeletal muscles. Computer Methods in Applied Mechanics and Engineering 151 , 419-433. DOI: 10.1016/S0045-7825(97)00162-X ;
Martins, J. A. C. and Pato, M. P. M. and Pires, E. B. (2006), A finite element model of skeletal muscles. Virtual and Physical Prototyping 1 , 159-170. DOI: 10.1080/17452750601040626 ;
Pandy, M. G. and Zajac, F. E. and Sim, E. and Levine, W. S. (1990), An optimal control model for maximum-height human jumping. Journal of Biomechanics 23 , 1185-1198. DOI: 10.1016/0021-9290(90)90376-E ;
T.J.R. Hughes (2000), The Finite Element Method: Linear Static and Dynamic Finite Element Analysis, Dover. ISBN: 978-0486411811
Results
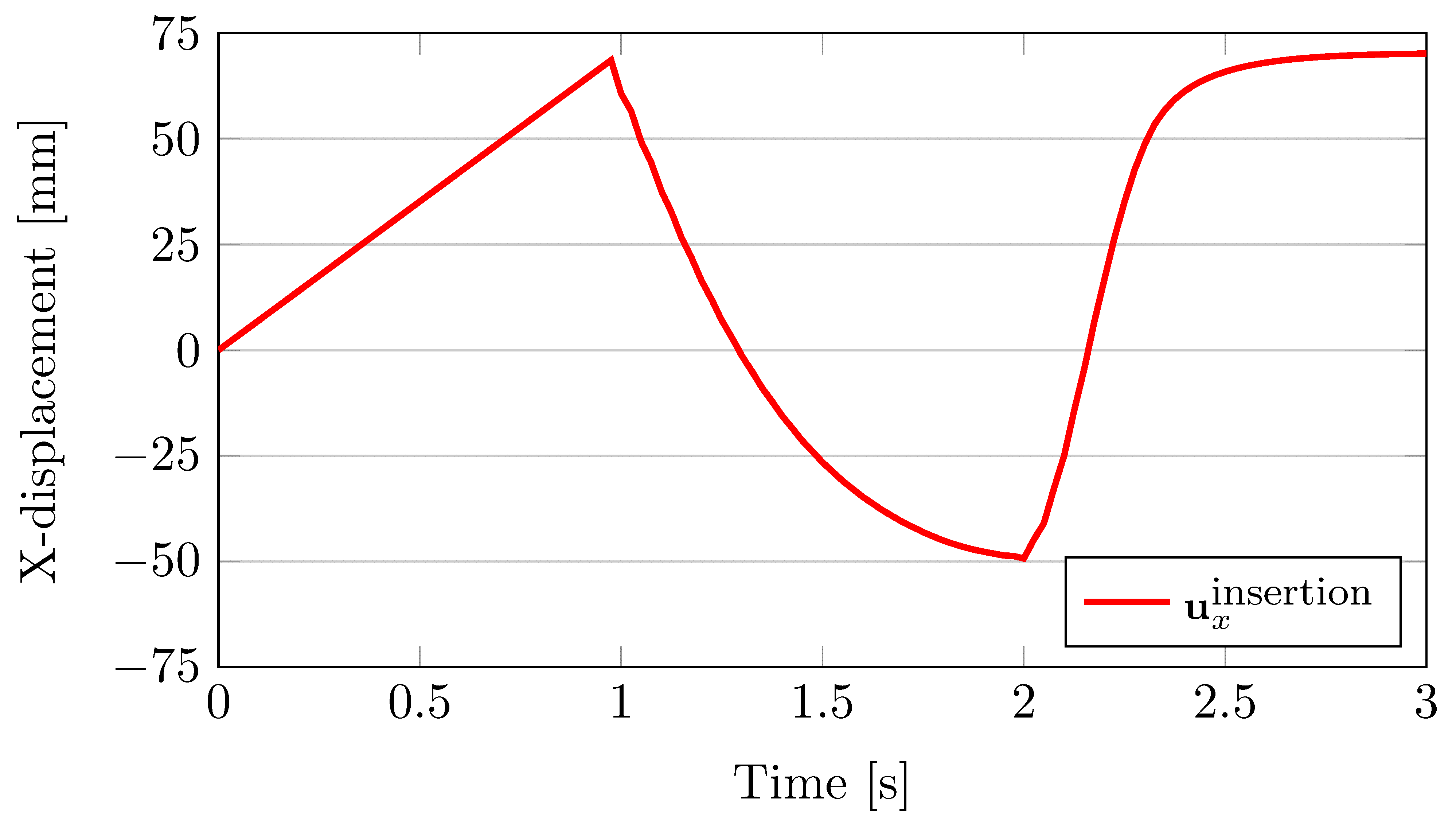
The displacement of the central point on the insertion surface (i.e. the traction boundary) is plotted against the simulation time when the muscle is made to undergo concentric contraction. Within the first second, when the muscle is completely passive, the displacement increases linearly due to the applied pressure that ramps to a maximum over this duration. This response is not entirely unsurprising for this geometrically symmetric, linear elastic body. When the muscle is activated, it shortens considerably until during the 1s for which the neural signal is applied. The activation level increases exponentially until is saturates near the 2s mark. At this point the neural signal is removed and the muscle starts to relax. The contractile level decreases exponentially and the muscle is nearly completely relaxed by the end of the simulation.
As a supplement to the above, the following animation shows the concentric contraction (under the assumption that it experiences no additional gravitational loading is present). All of the highlights that are discussed above can be observed in the gross displacement of the body, as well as the activation level that is visualised through the depiction of the underlying microstructure directions. This also shows how the muscle's cross-section influences the shortening along the length of the muscle.
Influence of gravity
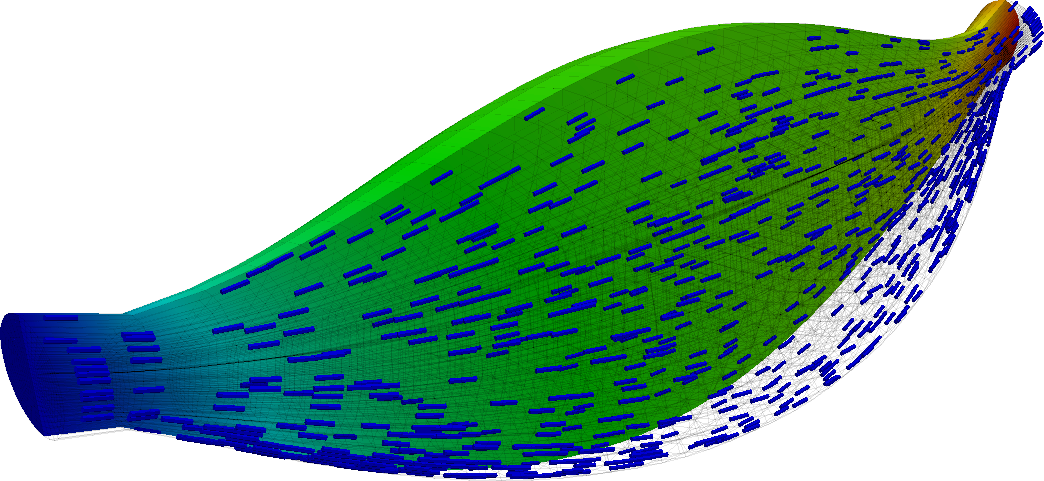
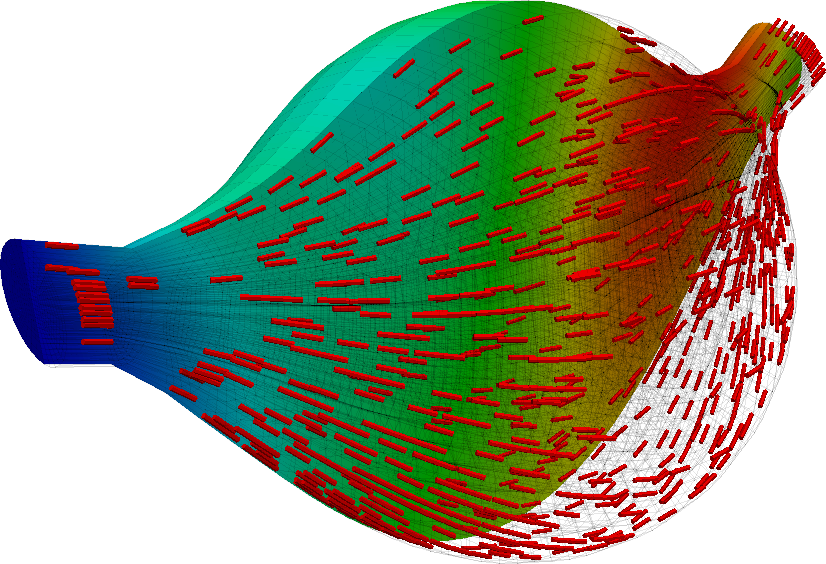
Just for fun, we can repeat the above experiment with a fraction of the full gravitational loading applied in the transverse direction. We apply only a fraction of the full load because the muscle is not sufficiently well constrained and does not see the support of its surrounding anatomy. The loading condition is thus somewhat unphysical and, due to the lack of constraint, the application of the full load results in excessive deformation.
Here we see the fully passive muscle with partial gravitational loading and a full traction load
and its counterpart solution when in the active stage.
The asymmetry of the solution is clearly observed, although the length change that it exhibits curing the concentric contraction cycle remains somewhat similar to that demonstrated before.
Annotated version of Linear_active_muscle_model.cc
/ * ---------------------------------------------------------------------
* Copyright (C) 2017 by the deal.II authors and
* Jean-Paul Pelteret and Tim Hamann
*
* This file is part of the deal.II library.
*
* The deal.II library is
free software; you can use it, redistribute
* it, and/or modify it under the terms of the GNU Lesser General
* Public License as published by the Free Software Foundation; either
* version 2.1 of the License, or (at your option) any later version.
* The full text of the license can be found in the file LICENSE at
* the top level of the deal.II distribution.
*
* ---------------------------------------------------------------------
* /
/ *
* Authors: Jean-Paul Pelteret, Tim Hamann,
* University of Erlangen-Nuremberg, 2017
*
* The support of this work by the European Research Council (ERC) through
* the Advanced Grant 289049 MOCOPOLY is gratefully acknowledged by the
* first author.
* /
Include files
#include <deal.II/base/quadrature_lib.h>
#include <deal.II/base/function.h>
#include <deal.II/base/logstream.h>
#include <deal.II/base/parameter_handler.h>
#include <deal.II/lac/vector.h>
#include <deal.II/lac/full_matrix.h>
#include <deal.II/lac/sparse_matrix.h>
#include <deal.II/lac/solver_cg.h>
#include <deal.II/lac/precondition.h>
#include <deal.II/lac/constraint_matrix.h>
#include <deal.II/grid/grid_out.h>
#include <deal.II/grid/manifold_lib.h>
#include <deal.II/grid/tria.h>
#include <deal.II/grid/grid_generator.h>
#include <deal.II/grid/grid_refinement.h>
#include <deal.II/grid/grid_tools.h>
#include <deal.II/grid/tria_accessor.h>
#include <deal.II/grid/tria_iterator.h>
#include <deal.II/grid/tria_boundary_lib.h>
#include <deal.II/dofs/dof_handler.h>
#include <deal.II/dofs/dof_accessor.h>
#include <deal.II/dofs/dof_tools.h>
#include <deal.II/fe/fe_values.h>
#include <deal.II/fe/fe_system.h>
#include <deal.II/fe/fe_q.h>
#include <deal.II/numerics/vector_tools.h>
#include <deal.II/numerics/matrix_tools.h>
#include <deal.II/numerics/data_out.h>
#include <deal.II/numerics/error_estimator.h>
#include <deal.II/physics/transformations.h>
#include <fstream>
#include <iostream>
#include <vector>
namespace LMM
{
Run-time parameters
There are several parameters that can be set in the code so we set up a ParameterHandler object to read in the choices at run-time.
Finite Element system
Here we specify the polynomial order used to approximate the solution. The quadrature order should be adjusted accordingly.
{
unsigned int poly_degree;
unsigned int quad_order;
static void
void
};
{
{
"Displacement system polynomial order" );
"Gauss quadrature order" );
}
}
{
{
}
}
Problem
Choose which problem is going to be solved
struct Problem
{
std::string problem;
static void
void
};
{
{
"The problem that is to be solved" );
}
}
{
{
problem = prm.
get (
"Problem" );
}
}
IsotonicContractionGeometry
Make adjustments to the geometry and discretisation of the isotonic contraction model from Martins2006.
struct IsotonicContraction
{
const double half_length_x = 10
e -3/2.0;
const double half_length_y = 10
e -3/2.0;
const double half_length_z = 1
e -3/2.0;
static void
void
};
{
}
{
}
BicepsBrachiiGeometry
Make adjustments to the geometry and discretisation of the biceps model.
struct BicepsBrachii
{
double axial_length;
double radius_insertion_origin;
double radius_midpoint;
unsigned int elements_along_axis;
unsigned int n_refinements_radial;
bool include_gravity;
double axial_force;
static void
void
};
{
{
"Axial length of the muscle" );
"Insertion and origin radius" );
"Radius at the midpoint of the muscle" );
"Global grid scaling factor" );
"Number of elements along the muscle axis" );
"Control the discretisation in the radial direction" );
"Include the effects of gravity (in the y-direction; "
" perpendicular to the muscle axis)" );
"Applied distributed axial force (in Newtons)" );
}
}
{
{
radius_insertion_origin = prm.
get_double (
"Radius insertion and origin" );
radius_midpoint = prm.
get_double (
"Radius midpoint" );
elements_along_axis = prm.
get_integer (
"Elements along axis" );
n_refinements_radial = prm.
get_integer (
"Radial refinements" );
include_gravity = prm.
get_bool (
"Gravity" );
}
AssertThrow (radius_midpoint >= radius_insertion_origin,
}
Neurological signal
struct NeurologicalSignal
{
double neural_signal_start_time;
double neural_signal_end_time;
static void
void
};
{
{
"Time at which to start muscle activation" );
"Time at which to remove muscle activation signal" );
}
}
{
{
neural_signal_start_time = prm.
get_double (
"Start time" );
neural_signal_end_time = prm.
get_double (
"End time" );
}
Assert (neural_signal_start_time < neural_signal_end_time,
}
Time
Set the timestep size \varDelta t and the simulation end-time.
struct Time
{
double delta_t;
double end_time;
double end_ramp_time;
static void
void
};
{
{
"End time" );
"Force ramp end time" );
"Time step size" );
}
}
{
{
}
}
All parameters
Finally we consolidate all of the above structures into a single container that holds all of our run-time selections.
public Problem,
public IsotonicContraction,
public BicepsBrachii,
public NeurologicalSignal,
public Time
{
AllParameters(const std::string &input_file);
static void
void
};
AllParameters::AllParameters(const std::string &input_file)
{
declare_parameters(prm);
parse_parameters(prm);
}
{
FESystem::declare_parameters(prm);
Problem::declare_parameters(prm);
IsotonicContraction::declare_parameters(prm);
BicepsBrachii::declare_parameters(prm);
NeurologicalSignal::declare_parameters(prm);
Time::declare_parameters(prm);
}
{
FESystem::parse_parameters(prm);
Problem::parse_parameters(prm);
IsotonicContraction::parse_parameters(prm);
BicepsBrachii::parse_parameters(prm);
NeurologicalSignal::parse_parameters(prm);
Time::parse_parameters(prm);
Override time setting for test defined in the literature
if (problem == "IsotonicContraction" )
{
end_time = 3.0;
end_ramp_time = 1.0;
delta_t = 0.1;
neural_signal_start_time = 1.0;
neural_signal_end_time = 2.0;
}
}
}
Body force values
template <int dim>
{
public :
BodyForce (const double rho,
virtual ~BodyForce () {}
const double rho;
const double g;
};
template <int dim>
BodyForce<dim>::BodyForce (const double rho,
:
rho (rho),
g (9.81),
M (direction)
{
}
template <int dim>
inline
void BodyForce<dim>::vector_value (
const Point<dim> &/ *p* /,
for (
unsigned int d=0;
d <dim; ++
d )
{
}
}
template <int dim>
void BodyForce<dim>::vector_value_list (
const std::vector<
Point<dim> > &points,
Assert (value_list.size() == points.size(),
const unsigned int n_points = points.size();
for (unsigned int p=0; p<n_points; ++p)
BodyForce<dim>::vector_value (points[p],
value_list[p]);
}
template <int dim>
{
public :
Traction (const double force,
const double area);
virtual ~Traction () {}
virtual void vector_value_list (
const std::vector<
Point<dim> > &points,
const double t;
};
template <int dim>
Traction<dim>::Traction (const double force,
const double area)
:
t (force/area)
{}
template <int dim>
inline
void Traction<dim>::vector_value (
const Point<dim> &/ *p* /,
Assume uniform distributed load
values(0) = t;
values(1) = 0.0;
values(2) = 0.0;
}
template <int dim>
void Traction<dim>::vector_value_list (
const std::vector<
Point<dim> > &points,
Assert (value_list.size() == points.size(),
const unsigned int n_points = points.size();
for (unsigned int p=0; p<n_points; ++p)
Traction<dim>::vector_value (points[p],
value_list[p]);
}
Utility functions
template <int dim>
inline
{
for (unsigned int i=0; i<dim; ++i)
for (unsigned int j=0; j<dim; ++j)
F[i][j] += grad[i][j];
}
template <int dim>
inline
{
for (unsigned int i=0; i<dim; ++i)
strain[i][i] = grad[i][i];
for (unsigned int i=0; i<dim; ++i)
for (unsigned int j=i+1; j<dim; ++j)
strain[i][j] = (grad[i][j] + grad[j][i]) / 2;
return strain;
}
Properties for muscle matrix
struct MuscleMatrix
{
static const double nu;
static const double lambda;
};
const double MuscleMatrix::E = 26e3;
const double MuscleMatrix::nu = 0.45;
const double MuscleMatrix::mu = MuscleMatrix::E/(2.0*(1.0 + MuscleMatrix::nu));
const double MuscleMatrix::lambda = 2.0*MuscleMatrix::mu *MuscleMatrix::nu/(1.0 - 2.0*MuscleMatrix::nu);
Local data for muscle fibres
#define convert_gf_to_N 1.0/101.97
#define convert_gf_per_cm2_to_N_per_m2 convert_gf_to_N*1e2*1e2
#define T0 6280.0*convert_gf_per_cm2_to_N_per_m2
A struct that governs the functioning of a single muscle fibre
template <int dim>
struct MuscleFibre
{
MuscleFibre (void )
: alpha (0.0),
alpha_t1 (0.0),
epsilon_f (0.0),
epsilon_c (0.0),
epsilon_c_t1 (0.0),
epsilon_c_dot (0.0)
{
}
: M (direction),
alpha (0.0),
alpha_t1 (0.0),
epsilon_f (0.0),
epsilon_c (0.0),
epsilon_c_t1 (0.0),
epsilon_c_dot (0.0)
{
ExcMessage (
"Fibre direction is not a unit vector" ));
}
void update_alpha (const double u,
const double dt);
const double dt);
{
return M;
}
double get_m_p () const ;
double get_m_s () const ;
double get_beta (const double dt) const ;
double get_gamma (const double dt) const ;
Postprocessing
const double &get_alpha() const
{
return alpha;
}
const double &get_epsilon_f() const
{
return epsilon_f;
}
const double &get_epsilon_c() const
{
return epsilon_c;
}
const double &get_epsilon_c_dot() const
{
return epsilon_c_dot;
}
private :
double alpha;
double alpha_t1;
double epsilon_f;
double epsilon_c;
double epsilon_c_t1;
double epsilon_c_dot;
double get_f_c_L () const ;
double get_m_c_V () const ;
double get_c_c_V () const ;
};
template <int dim>
void MuscleFibre<dim>::update_alpha (const double u,
const double dt)
{
static const double tau_r = 0.15;
static const double tau_f = 0.15;
static const double alpha_min = 0;
if (u == 1.0)
alpha = (alpha_t1*tau_r*tau_f + dt*tau_f) / (tau_r*tau_f + dt*tau_f);
else if (u == 0)
alpha = (alpha_t1*tau_r*tau_f + dt*alpha_min*tau_r) / (tau_r*tau_f + dt*tau_r);
else
{
const double b = 1.0/tau_r - 1.0/tau_f;
const double c = 1.0/tau_f;
const double d = alpha_min/tau_f;
const double f1 = 1.0/tau_r - alpha_min/tau_f;
const double p =
b *u + c;
const double q = f1*u +
d ;
alpha = (q*dt + alpha_t1)/(1.0 + p*dt);
}
}
template <int dim>
double MuscleFibre<dim>::get_m_p () const
static const double A = 8.568e-4*convert_gf_per_cm2_to_N_per_m2;
static const double a = 12.43;
if (epsilon_f >= 0.0)
{
100 times more compliant than Martins2006
static const double m_p = 2.0*A*a/1e2;
return m_p;
}
else
return 0.0;
}
template <int dim>
double MuscleFibre<dim>::get_m_s (void ) const
const double epsilon_s = epsilon_f - epsilon_c;
if (epsilon_s >= -1e-6)
return 10.0;
else
return 0.0;
}
template <int dim>
double MuscleFibre<dim>::get_f_c_L (void ) const
if (epsilon_c <= 0.5 && epsilon_c >= -0.5)
return 1.0;
else
return 0.0;
}
template <int dim>
double MuscleFibre<dim>::get_m_c_V (void ) const
if (epsilon_c_dot < -5.0)
return 0.0;
else if (epsilon_c_dot <= 3.0)
return 1.0/5.0;
else
return 0.0;
}
template <int dim>
double MuscleFibre<dim>::get_c_c_V (void ) const
if (epsilon_c_dot < -5.0)
return 0.0;
else if (epsilon_c_dot <= 3.0)
return 1.0;
else
return 1.6;
}
template <int dim>
double MuscleFibre<dim>::get_beta(const double dt) const
return get_f_c_L()*get_m_c_V()*alpha/dt + get_m_s();
}
template <int dim>
double MuscleFibre<dim>::get_gamma(const double dt) const
return get_f_c_L()*alpha*(get_m_c_V()*epsilon_c_t1/dt - get_c_c_V());
}
template <int dim>
const double dt)
{
Values from previous state These were the values that were used in the assembly, so we must use them in the update step to be consistant. Need to compute these before we overwrite epsilon_c_t1
const double m_s = get_m_s();
const double beta = get_beta(dt);
const double gamma = get_gamma(dt);
Update current state
alpha_t1 = alpha;
epsilon_c_t1 = epsilon_c;
epsilon_c = (m_s*epsilon_f + gamma)/beta;
epsilon_c_dot = (epsilon_c - epsilon_c_t1)/dt;
}
The LinearMuscleModelProblem class template
template <int dim>
class LinearMuscleModelProblem
{
public :
LinearMuscleModelProblem (const std::string &input_file);
~LinearMuscleModelProblem ();
private :
void make_grid ();
void setup_muscle_fibres ();
double get_neural_signal (const double time);
void update_fibre_activation (const double time);
void update_fibre_state ();
void setup_system ();
void assemble_system (const double time);
void apply_boundary_conditions ();
void solve ();
void output_results (const unsigned int timestep,
const double time) const ;
Parameters::AllParameters parameters;
Time
const double t_end;
const double dt;
const double t_ramp_end;
Loading
const BodyForce<dim> body_force;
const Traction<dim> traction;
Local data
std::vector< std::vector<MuscleFibre<dim> > > fibre_data;
Constitutive functions for assembly
const unsigned int q_point_cell) const ;
const unsigned int q_point_cell) const ;
};
LinearMuscleModelProblem::LinearMuscleModelProblem
template <int dim>
LinearMuscleModelProblem<dim>::LinearMuscleModelProblem (const std::string &input_file)
:
parameters(input_file),
dof_handler (triangulation),
fe (
FE_Q <dim>(parameters.poly_degree), dim),
qf_cell (parameters.quad_order),
qf_face (parameters.quad_order),
t_end (parameters.end_time),
dt (parameters.delta_t),
t_ramp_end(parameters.end_ramp_time),
body_force ((parameters.problem == "BicepsBrachii" &¶meters.include_gravity == true) ?
BodyForce<dim>(0.375*1000.0,
Tensor <1,dim>({0,-1,0})) :
BodyForce<dim>(0.0,
Tensor <1,dim>({0,0,1})) ),
traction (parameters.problem == "BicepsBrachii" ?
Traction<dim>(parameters.axial_force,
M_PI*std::pow(parameters.radius_insertion_origin *parameters.scale,2.0) ) :
Traction<dim>(4.9*convert_gf_to_N,
(2.0*parameters.half_length_y)*(2.0*parameters.half_length_z)) )
{
}
LinearMuscleModelProblem::~LinearMuscleModelProblem
template <int dim>
LinearMuscleModelProblem<dim>::~LinearMuscleModelProblem ()
{
}
LinearMuscleModelProblem::make_grid
template <int dim>
struct BicepsGeometry
{
BicepsGeometry(const double axial_length,
const double radius_ins_orig,
const double radius_mid)
:
ax_lgth (axial_length),
r_ins_orig (radius_ins_orig),
r_mid (radius_mid)
{}
The radial profile of the muscle This provides the new coordinates for points pt on a cylinder of radius r_ins_orig and length ax_lgth to be moved to in order to create the physiologically representative geometry of the muscle
ExcMessage (
"All points must have x-coordinate > 0" ));
const double r_scale = get_radial_scaling_factor(pt_0[0]);
return pt_0 +
Point<dim> (0.0, r_scale*pt_0[1], r_scale*pt_0[2]);
}
return profile(pt);
}
Provides the muscle direction at the point pt in the real geometry (one that has undergone the transformation given by the profile() function) and subequent grid rescaling. The directions are given by the gradient of the transformation function (i.e. the fibres are orientated by the curvature of the muscle).
So, being lazy, we transform the current point back to the original point on the completely unscaled cylindrical grid. We then evaluate the transformation at two points (axially displaced) very close to the point of interest. The normalised vector joining the transformed counterparts of the perturbed points is the gradient of the transformation function and, thus, defines the fibre direction.
const double &grid_scale) const
{
static const double eps = 1
e -6;
static const double tol = 1
e -9;
return dir;
}
private :
const double ax_lgth;
const double r_ins_orig;
const double r_mid;
double get_radial_scaling_factor (const double &x) const
{
Expect all grid points with X>=0, but we provide a tolerant location for points "on" the Cartesian plane X=0
const double lgth_frac = std::max(x/ax_lgth,0.0);
const double amplitude = 0.25*(r_mid - r_ins_orig);
const double phase_shift = M_PI;
const double y_shift = 1.0;
const double wave_func = y_shift + std::cos(phase_shift + 2.0*M_PI*lgth_frac);
return std::sqrt(amplitude*wave_func);
}
{
ExcMessage (
"All points must have x-coordinate > 0" ));
const double r_scale = get_radial_scaling_factor(pt[0]);
const double trans_inv_scale = 1.0/(1.0+r_scale);
return Point<dim> (pt[0], trans_inv_scale*pt[1], trans_inv_scale*pt[2]);
}
};
template <int dim>
void LinearMuscleModelProblem<dim>::make_grid ()
{
if (parameters.problem == "IsotonicContraction" )
{
-parameters.half_length_y,
-parameters.half_length_z);
parameters.half_length_y,
parameters.half_length_z);
triangulation.begin_active(), endc = triangulation.end();
for (; cell != endc; ++cell)
{
for (unsigned int face = 0; face < GeometryInfo<dim>::faces_per_cell; ++face)
{
if (cell->face(face)->at_boundary() == true )
{
if (cell->face(face)->center()[0] == -parameters.half_length_x)
cell->face(face)->set_boundary_id(parameters.bid_CC_dirichlet_symm_X);
else if (cell->face(face)->center()[0] == parameters.half_length_x)
cell->face(face)->set_boundary_id(parameters.bid_CC_neumann);
else if (std::abs(cell->face(face)->center()[2]) == parameters.half_length_z)
cell->face(face)->set_boundary_id(parameters.bid_CC_dirichlet_symm_Z);
}
}
}
triangulation.refine_global (1);
}
else if (parameters.problem == "BicepsBrachii" )
{
parameters.radius_insertion_origin);
cell != tria_cap.
end (); ++cell)
{
for (unsigned int face = 0; face < GeometryInfo<2>::faces_per_cell; ++face)
{
if (cell->face(face)->at_boundary() == true )
}
}
parameters.elements_along_axis,
parameters.axial_length,
triangulation);
struct GridRotate
{
{
}
};
Rotate grid so that the length is axially coincident and aligned with the X-axis
Deform the grid into something that vaguely resemble's a Biceps Brachii
parameters.radius_insertion_origin,
parameters.radius_midpoint), triangulation);
Set boundary IDs
triangulation.begin_active(), endc = triangulation.end();
for (; cell != endc; ++cell)
{
for (unsigned int face = 0; face < GeometryInfo<dim>::faces_per_cell; ++face)
{
if (cell->face(face)->at_boundary() == true )
{
static const double tol =1
e -6;
if (std::abs(cell->face(face)->center()[0]) < tol)
cell->face(face)->set_boundary_id(parameters.bid_BB_dirichlet_X);
else if (std::abs(cell->face(face)->center()[0] - parameters.axial_length) < tol)
cell->face(face)->set_boundary_id(parameters.bid_BB_neumann);
}
}
}
Finally resize the grid
LinearMuscleModelProblem::setup_muscle_fibres
template <int dim>
void LinearMuscleModelProblem<dim>::setup_muscle_fibres ()
{
fibre_data.clear();
const unsigned int n_cells = triangulation.n_active_cells();
fibre_data.resize(n_cells);
const unsigned int n_q_points_cell = qf_cell.
size ();
if (parameters.problem == "IsotonicContraction" )
{
for (unsigned int cell_no=0; cell_no<triangulation.n_active_cells(); ++cell_no)
{
fibre_data[cell_no].resize(n_q_points_cell);
for (unsigned int q_point_cell=0; q_point_cell<n_q_points_cell; ++q_point_cell)
{
fibre_data[cell_no][q_point_cell] = fibre_template;
}
}
}
else if (parameters.problem == "BicepsBrachii" )
{
BicepsGeometry<dim> bicep_geom (parameters.axial_length,
parameters.radius_insertion_origin,
parameters.radius_midpoint);
unsigned int cell_no = 0;
cell = triangulation.begin_active();
cell != triangulation.end();
++cell, ++cell_no)
{
Assert (cell_no<fibre_data.size(),
ExcMessage (
"Trying to access fibre data not stored for this cell index" ));
fibre_data[cell_no].resize(n_q_points_cell);
for (unsigned int q_point_cell=0; q_point_cell<n_q_points_cell; ++q_point_cell)
{
fibre_data[cell_no][q_point_cell] = MuscleFibre<dim>(bicep_geom.direction(pt,parameters.scale));
}
}
}
else
}
LinearMuscleModelProblem::update_fibre_state
template <int dim>
double LinearMuscleModelProblem<dim>::get_neural_signal (const double time)
{
Note: 40 times less force generated than Martins2006 This is necessary due to the (compliant) linear tissue model
return (time > parameters.neural_signal_start_time && time < parameters.neural_signal_end_time ?
1.0/40.0 :
0.0);
}
template <int dim>
void LinearMuscleModelProblem<dim>::update_fibre_activation (const double time)
{
const double u = get_neural_signal(time);
const unsigned int n_q_points_cell = qf_cell.
size ();
for (unsigned int cell=0; cell<triangulation.n_active_cells(); ++cell)
{
for (unsigned int q_point_cell=0; q_point_cell<n_q_points_cell; ++q_point_cell)
{
MuscleFibre<dim> &fibre = fibre_data[cell][q_point_cell];
fibre.update_alpha(u,dt);
}
}
}
template <int dim>
void LinearMuscleModelProblem<dim>::update_fibre_state ()
{
const unsigned int n_q_points_cell = qf_cell.
size ();
Displacement gradient
std::vector< std::vector< Tensor<1,dim> > > u_grads (n_q_points_cell,
unsigned int cell_no = 0;
cell!=dof_handler.
end (); ++cell, ++cell_no)
{
Assert (cell_no<fibre_data.size(),
ExcMessage (
"Trying to access fibre data not stored for this cell index" ));
for (unsigned int q_point_cell=0; q_point_cell<n_q_points_cell; ++q_point_cell)
{
Assert (q_point_cell<fibre_data[cell_no].size(),
ExcMessage (
"Trying to access fibre data not stored for this cell and qp index" ));
MuscleFibre<dim> &fibre = fibre_data[cell_no][q_point_cell];
fibre.update_state(strain_tensor, dt);
}
}
}
LinearMuscleModelProblem::setup_system
template <int dim>
void LinearMuscleModelProblem<dim>::setup_system ()
{
hanging_node_constraints.clear ();
hanging_node_constraints);
hanging_node_constraints.close ();
hanging_node_constraints.condense (sparsity_pattern);
system_matrix.reinit (sparsity_pattern);
std::cout << " Number of active cells: "
<< triangulation.n_active_cells()
<< std::endl;
std::cout << " Number of degrees of freedom: "
<< std::endl;
}
LinearMuscleModelProblem::assemble_system
template <int dim>
LinearMuscleModelProblem<dim>::get_stiffness_tensor (const unsigned int cell,
const unsigned int q_point_cell) const
Assert (cell<fibre_data.size(),
ExcMessage (
"Trying to access fibre data not stored for this cell index" ));
Assert (q_point_cell<fibre_data[cell].size(),
ExcMessage (
"Trying to access fibre data not stored for this cell and qp index" ));
const MuscleFibre<dim> &fibre = fibre_data[cell][q_point_cell];
Matrix
const double lambda = MuscleMatrix::lambda;
const double mu = MuscleMatrix::mu;
Fibre
const double m_p = fibre.get_m_p();
const double m_s = fibre.get_m_s();
const double beta = fibre.get_beta(dt);
const double Cf = T0*(m_p + m_s*(1.0 - m_s/beta));
for (unsigned int i=0; i < dim; ++i)
for (unsigned int j=i; j < dim; ++j)
for (unsigned int k=0; k < dim; ++k)
for (
unsigned int l=k;
l < dim; ++
l )
{
Matrix contribution
C [i][j][k][
l ] = lambda * I[i][j]*I[k][
l ]
+
mu * (I[i][k]*I[j][
l ] + I[i][
l ]*I[j][k]);
Fibre contribution (Passive + active branches)
C [i][j][k][
l ] += Cf * M[i]*M[j]*M[k]*M[
l ];
}
}
template <int dim>
LinearMuscleModelProblem<dim>::get_rhs_tensor (const unsigned int cell,
const unsigned int q_point_cell) const
Assert (cell<fibre_data.size(),
ExcMessage (
"Trying to access fibre data not stored for this cell index" ));
Assert (q_point_cell<fibre_data[cell].size(),
ExcMessage (
"Trying to access fibre data not stored for this cell and qp index" ));
const MuscleFibre<dim> &fibre = fibre_data[cell][q_point_cell];
const double m_s = fibre.get_m_s();
const double beta = fibre.get_beta(dt);
const double gamma = fibre.get_gamma(dt);
const double Sf = T0*(m_s*gamma/beta);
for (unsigned int i=0; i < dim; ++i)
for (unsigned int j=i; j < dim; ++j)
{
Fibre contribution (Active branch)
S[i][j] = Sf * M[i]*M[j];
}
return S;
}
LinearMuscleModelProblem::assemble_system
template <int dim>
void LinearMuscleModelProblem<dim>::assemble_system (const double time)
{
Reset system
system_matrix = 0;
system_rhs = 0;
const unsigned int n_q_points_cell = qf_cell.
size ();
const unsigned int n_q_points_face = qf_face.size();
std::vector<types::global_dof_index> local_dof_indices (dofs_per_cell);
Loading
std::vector<Vector<double> > body_force_values (n_q_points_cell,
std::vector<Vector<double> > traction_values (n_q_points_face,
unsigned int cell_no = 0;
cell!=dof_handler.
end (); ++cell, ++cell_no)
{
cell_rhs = 0;
body_force_values);
for (unsigned int q_point_cell=0; q_point_cell<n_q_points_cell; ++q_point_cell)
{
for (unsigned int I=0; I<dofs_per_cell; ++I)
{
const unsigned int
for (unsigned int J=0; J<dofs_per_cell; ++J)
{
const unsigned int
for (unsigned int k=0; k < dim; ++k)
for (
unsigned int l=0;
l < dim; ++
l )
C [component_I][k][component_J][
l ] *
fe_values.
JxW (q_point_cell);
}
}
for (unsigned int I=0; I<dofs_per_cell; ++I)
{
const unsigned int
cell_rhs(I)
body_force_values[q_point_cell](component_I) *
fe_values.
JxW (q_point_cell);
for (unsigned int k=0; k < dim; ++k)
cell_rhs(I)
R[component_I][k]) *
fe_values.
JxW (q_point_cell);
}
}
for (unsigned int face = 0; face <GeometryInfo<dim>::faces_per_cell; ++face)
{
if (cell->face(face)->at_boundary() == true &&
((parameters.problem == "IsotonicContraction" &&
cell->face(face)->boundary_id() == parameters.bid_CC_neumann) ||
(parameters.problem == "BicepsBrachii" &&
cell->face(face)->boundary_id() == parameters.bid_BB_neumann)) )
{
fe_face_values.reinit(cell, face);
traction_values);
Scale applied traction according to time
const double ramp = (time <= t_ramp_end ? time/t_ramp_end : 1.0);
for (unsigned int q_point_face = 0; q_point_face < n_q_points_face; ++q_point_face)
traction_values[q_point_face] *= ramp;
for (unsigned int q_point_face = 0; q_point_face < n_q_points_face; ++q_point_face)
{
for (unsigned int I=0; I<dofs_per_cell; ++I)
{
const unsigned int
cell_rhs(I)
traction_values[q_point_face][component_I]*
fe_face_values.
JxW (q_point_face);
}
}
}
}
cell->get_dof_indices (local_dof_indices);
for (unsigned int i=0; i<dofs_per_cell; ++i)
{
for (unsigned int j=0; j<dofs_per_cell; ++j)
system_matrix.add (local_dof_indices[i],
local_dof_indices[j],
system_rhs(local_dof_indices[i]) += cell_rhs(i);
}
}
hanging_node_constraints.condense (system_matrix);
hanging_node_constraints.condense (system_rhs);
}
template <int dim>
void LinearMuscleModelProblem<dim>::apply_boundary_conditions ()
{
std::map<types::global_dof_index,double> boundary_values;
if (parameters.problem == "IsotonicContraction" )
{
Symmetry condition on -X faces
{
component_mask_x.set(0, true );
parameters.bid_CC_dirichlet_symm_X,
boundary_values,
component_mask_x);
}
Symmetry condition on -Z/+Z faces
{
component_mask_z.set(2, true );
parameters.bid_CC_dirichlet_symm_Z,
boundary_values,
component_mask_z);
}
Fixed point on -X face
{
const Point<dim> fixed_point (-parameters.half_length_x,0.0,0.0);
std::vector<types::global_dof_index> fixed_dof_indices;
bool found_point_of_interest = false ;
endc = dof_handler.
end (); cell != endc; ++cell)
{
for (unsigned int face = 0; face < GeometryInfo<dim>::faces_per_cell; ++face)
{
We know that the fixed point is on the -X Dirichlet boundary
if (cell->face(face)->at_boundary() == true &&
cell->face(face)->boundary_id() == parameters.bid_CC_dirichlet_symm_X)
{
for (unsigned int face_vertex_index = 0; face_vertex_index < GeometryInfo<dim>::vertices_per_face; ++face_vertex_index)
{
if (cell->face(face)->vertex(face_vertex_index).distance(fixed_point) < 1
e -6)
{
found_point_of_interest = true ;
for (unsigned int index_component = 0; index_component < dim; ++index_component)
fixed_dof_indices.push_back(cell->face(face)->vertex_dof_index(face_vertex_index,
index_component));
}
if (found_point_of_interest == true ) break ;
}
}
if (found_point_of_interest == true ) break ;
}
if (found_point_of_interest == true ) break ;
}
Assert (found_point_of_interest ==
true ,
ExcMessage (
"Didn't find point of interest" ));
AssertThrow (fixed_dof_indices.size() == dim,
ExcMessage (
"Didn't find the correct number of DoFs to fix" ));
for (unsigned int i=0; i < fixed_dof_indices.size(); ++i)
boundary_values[fixed_dof_indices[i]] = 0.0;
}
}
else if (parameters.problem == "BicepsBrachii" )
{
if (parameters.include_gravity == false )
{
Symmetry condition on -X surface
{
component_mask_x.set(0, true );
parameters.bid_BB_dirichlet_X,
boundary_values,
component_mask_x);
}
Fixed central point on -X surface
{
std::vector<types::global_dof_index> fixed_dof_indices;
bool found_point_of_interest = false ;
endc = dof_handler.
end (); cell != endc; ++cell)
{
for (unsigned int face = 0; face < GeometryInfo<dim>::faces_per_cell; ++face)
{
We know that the fixed point is on the -X Dirichlet boundary
if (cell->face(face)->at_boundary() == true &&
cell->face(face)->boundary_id() == parameters.bid_BB_dirichlet_X)
{
for (unsigned int face_vertex_index = 0; face_vertex_index < GeometryInfo<dim>::vertices_per_face; ++face_vertex_index)
{
if (cell->face(face)->vertex(face_vertex_index).distance(fixed_point) < 1
e -6)
{
found_point_of_interest = true ;
for (unsigned int index_component = 0; index_component < dim; ++index_component)
fixed_dof_indices.push_back(cell->face(face)->vertex_dof_index(face_vertex_index,
index_component));
}
if (found_point_of_interest == true ) break ;
}
}
if (found_point_of_interest == true ) break ;
}
if (found_point_of_interest == true ) break ;
}
Assert (found_point_of_interest ==
true ,
ExcMessage (
"Didn't find point of interest" ));
AssertThrow (fixed_dof_indices.size() == dim,
ExcMessage (
"Didn't find the correct number of DoFs to fix" ));
for (unsigned int i=0; i < fixed_dof_indices.size(); ++i)
boundary_values[fixed_dof_indices[i]] = 0.0;
}
}
else
{
When we apply gravity, some additional constraints are required to support the load of the muscle, as the material response is more compliant than would be the case in reality.
Symmetry condition on -X surface
{
parameters.bid_BB_dirichlet_X,
boundary_values,
component_mask_x);
}
Symmetry condition on -X surface
{
component_mask_x.set(1, true );
component_mask_x.set(2, true );
parameters.bid_BB_neumann,
boundary_values,
component_mask_x);
}
}
Roller condition at central point on +X face
{
const Point<dim> roller_point (parameters.axial_length*parameters.scale,0.0,0.0);
std::vector<types::global_dof_index> fixed_dof_indices;
bool found_point_of_interest = false ;
endc = dof_handler.
end (); cell != endc; ++cell)
{
for (unsigned int face = 0; face < GeometryInfo<dim>::faces_per_cell; ++face)
{
We know that the fixed point is on the +X Neumann boundary
if (cell->face(face)->at_boundary() == true &&
cell->face(face)->boundary_id() == parameters.bid_BB_neumann)
{
for (unsigned int face_vertex_index = 0; face_vertex_index < GeometryInfo<dim>::vertices_per_face; ++face_vertex_index)
{
if (cell->face(face)->vertex(face_vertex_index).distance(roller_point) < 1
e -6)
{
found_point_of_interest = true ;
for (unsigned int index_component = 1; index_component < dim; ++index_component)
fixed_dof_indices.push_back(cell->face(face)->vertex_dof_index(face_vertex_index,
index_component));
}
if (found_point_of_interest == true ) break ;
}
}
if (found_point_of_interest == true ) break ;
}
if (found_point_of_interest == true ) break ;
}
Assert (found_point_of_interest ==
true ,
ExcMessage (
"Didn't find point of interest" ));
AssertThrow (fixed_dof_indices.size() == dim-1,
ExcMessage (
"Didn't find the correct number of DoFs to fix" ));
for (unsigned int i=0; i < fixed_dof_indices.size(); ++i)
boundary_values[fixed_dof_indices[i]] = 0.0;
}
}
else
system_matrix,
solution,
system_rhs);
}
LinearMuscleModelProblem::solve
template <int dim>
void LinearMuscleModelProblem<dim>::solve ()
{
cg.solve (system_matrix, solution, system_rhs,
preconditioner);
hanging_node_constraints.distribute (solution);
}
LinearMuscleModelProblem::output_results
template <int dim>
void LinearMuscleModelProblem<dim>::output_results (const unsigned int timestep,
const double time) const
Visual output: FEM results
{
std::string filename = "solution-" ;
filename += ".vtk" ;
std::ofstream output (filename.c_str());
std::vector<DataComponentInterpretation::DataComponentInterpretation>
data_component_interpretation(dim,
std::vector<std::string> solution_name(dim, "displacement" );
data_out.add_data_vector (solution, solution_name,
data_component_interpretation);
data_out.build_patches ();
data_out.write_vtk (output);
}
Visual output: FEM data
{
std::string filename = "fibres-" ;
filename += ".vtk" ;
std::ofstream output (filename.c_str());
output
<< "# vtk DataFile Version 3.0" << std::endl
<< "# " << std::endl
<< "ASCII" << std::endl
<< "DATASET POLYDATA" << std::endl << std::endl;
Extract fibre data from quadrature points
const unsigned int n_cells = triangulation.n_active_cells();
const unsigned int n_q_points_cell = qf_cell.
size ();
Data that we'll be outputting
std::vector<std::string> results_fibre_names;
results_fibre_names.push_back("alpha" );
results_fibre_names.push_back("epsilon_f" );
results_fibre_names.push_back("epsilon_c" );
results_fibre_names.push_back("epsilon_c_dot" );
const unsigned int n_results = results_fibre_names.size();
const unsigned int n_data_points = n_cells*n_q_points_cell;
std::vector< Point<dim> > output_points(n_data_points);
std::vector< Tensor<1,dim> > output_displacements(n_data_points);
std::vector< Tensor<1,dim> > output_directions(n_data_points);
std::vector< std::vector<double> > output_values(n_results, std::vector<double>(n_data_points));
Displacement
std::vector< Vector<double> > u_values (n_q_points_cell,
Displacement gradient
std::vector< std::vector< Tensor<1,dim> > > u_grads (n_q_points_cell,
unsigned int cell_no = 0;
unsigned int fibre_no = 0;
cell != dof_handler.
end ();
++cell, ++cell_no)
{
for (unsigned int q_point_cell=0; q_point_cell<n_q_points_cell; ++q_point_cell, ++fibre_no)
{
const MuscleFibre<dim> &fibre = fibre_data[cell_no][q_point_cell];
for (
unsigned int d=0;
d <dim; ++
d )
output_displacements[fibre_no][d] = u_values[q_point_cell][d];
Direction (spatial configuration)
output_directions[fibre_no] = get_deformation_gradient(u_grads[q_point_cell])*fibre.get_M();
output_directions[fibre_no] /= output_directions[fibre_no].norm();
Fibre values
output_values[0][fibre_no] = fibre.get_alpha();
output_values[1][fibre_no] = fibre.get_epsilon_f();
output_values[2][fibre_no] = fibre.get_epsilon_c();
output_values[3][fibre_no] = fibre.get_epsilon_c_dot();
}
}
FIBRE POSITION
output
<< "POINTS "
<< n_data_points
<< " float" << std::endl;
for (unsigned int i=0; i < n_data_points; ++i)
{
for (unsigned int j=0; j < dim; ++j)
{
output << (output_points)[i][j] << "\t" ;
}
output << std::endl;
}
HEADER FOR POINT DATA
output << "\nPOINT_DATA "
<< n_data_points
<< std::endl << std::endl;
FIBRE DISPLACEMENTS
output
<< "VECTORS displacement float"
<< std::endl;
for (unsigned int i = 0; i < n_data_points; ++i)
{
for (unsigned int j=0; j < dim; ++j)
{
output << (output_displacements)[i][j] << "\t" ;
}
output << std::endl;
}
output << std::endl;
FIBRE DIRECTIONS
output
<< "VECTORS direction float"
<< std::endl;
for (unsigned int i = 0; i < n_data_points; ++i)
{
for (unsigned int j=0; j < dim; ++j)
{
output << (output_directions)[i][j] << "\t" ;
}
output << std::endl;
}
output << std::endl;
POINT DATA
for (unsigned int v=0; v < n_results; ++v)
{
output
<< "SCALARS "
<< results_fibre_names[v]
<< " float 1" << std::endl
<< "LOOKUP_TABLE default "
<< std::endl;
for (unsigned int i=0; i<n_data_points; ++i)
{
output << (output_values)[v][i] << " " ;
}
output << std::endl;
}
}
Output X-displacement at measured point
{
const Point<dim> meas_pt (parameters.problem ==
"IsotonicContraction" ?
Point<dim> (parameters.axial_length*parameters.scale, 0.0, 0.0) );
const unsigned int index_of_interest = 0;
bool found_point_of_interest = false ;
endc = dof_handler.
end (); cell != endc; ++cell)
{
for (unsigned int face = 0; face < GeometryInfo<dim>::faces_per_cell; ++face)
{
We know that the measurement point is on the Neumann boundary
if (cell->face(face)->at_boundary() == true &&
((parameters.problem == "IsotonicContraction" &&
cell->face(face)->boundary_id() == parameters.bid_CC_neumann) ||
(parameters.problem == "BicepsBrachii" &&
cell->face(face)->boundary_id() == parameters.bid_BB_neumann)) )
{
for (unsigned int face_vertex_index = 0; face_vertex_index < GeometryInfo<dim>::vertices_per_face; ++face_vertex_index)
{
if (cell->face(face)->vertex(face_vertex_index).distance(meas_pt) < 1
e -6)
{
found_point_of_interest = true ;
dof_of_interest = cell->face(face)->vertex_dof_index(face_vertex_index,
index_of_interest);
}
if (found_point_of_interest == true ) break ;
}
}
if (found_point_of_interest == true ) break ;
}
if (found_point_of_interest == true ) break ;
}
Assert (found_point_of_interest ==
true ,
ExcMessage (
"Didn't find point of interest" ));
const std::string filename = "displacement_POI.csv" ;
std::ofstream output;
if (timestep == 0)
{
output.open(filename.c_str(), std::ofstream::out);
output
<< "Time [s]" << "," << "X-displacement [mm]" << std::endl;
}
else
output.open(filename.c_str(), std::ios_base::app);
output
<< time
<< ","
<< solution[dof_of_interest]*1e3
<< std::endl;
}
}
LinearMuscleModelProblem::run
template <int dim>
void LinearMuscleModelProblem<dim>::run ()
{
make_grid();
setup_system ();
setup_muscle_fibres ();
const bool do_grid_refinement = false;
double time = 0.0;
for (unsigned int timestep=0; time<=t_end; ++timestep, time+=dt)
{
std::cout
<< "Timestep " << timestep
<< " @ time " << time
<< std::endl;
First we update the fibre activation level based on the current time
update_fibre_activation(time);
Next we assemble the system and enforce boundary conditions. Here we assume that the system and fibres have a fixed state, and we will assemble based on how epsilon_c will update given the current state of the body.
assemble_system (time);
apply_boundary_conditions ();
Then we solve the linear system
Now we update the fibre state based on the new displacement solution and the constitutive parameters assumed to govern the stiffness of the fibres at the previous state. i.e. We follow through with assumed update conditions used in the assembly phase.
Output some values to file
output_results (timestep, time);
}
}
}
The main function
int main ()
{
try
{
::deallog.depth_console (0);
const unsigned int dim = 3;
LMM::LinearMuscleModelProblem<dim> lmm_problem ("parameters.prm" );
lmm_problem.run ();
}
catch (std::exception &exc)
{
std::cerr << std::endl << std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Exception on processing: " << std::endl
<< exc.what() << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
catch (...)
{
std::cerr << std::endl << std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Unknown exception!" << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
return 0;
}







 1.8.14
1.8.14