This tutorial depends on step-15, step-37.
This program was contributed by Fabian Castelli.
A version of this code was presented and discussed in [30] G.F. Castelli: Numerical Investigation of Cahn-Hilliard-Type Phase-Field Models for Battery Active Particles, PhD thesis, Karlsruhe Institute of Technology (KIT), 2021. (To be published)
Fabian Castelli acknowledges financial support by the German Research Foundation (DFG) through the Research Training Group 2218 SiMET – Simulation of mechano-electro-thermal processes in lithium-ion batteries, project number 281041241.
Finally Fabian Castelli would like to thank Timo Heister for the encouragement and advice in writing this tutorial.
Introduction
The aim of this tutorial program is to demonstrate how to solve a nonlinear problem using Newton's method within the matrix-free framework. This tutorial combines several techniques already introduced in step-15, step-16, step-37, step-48 and others.
Problem formulation
On the unit circle \(\Omega = \bigl\{ x \in \mathbb{R}^2 : \|x\| \leq 1 \bigr\}\) we consider the following nonlinear elliptic boundary value problem subject to a homogeneous Dirichlet boundary condition: Find a function \(u\colon\Omega\to\mathbb{R}\) such that it holds:
\begin{align*}
- \Delta u &= \exp(u) & \quad & \text{in } \Omega,\\
u &= 0 & \quad & \text{on } \partial\Omega.
\end{align*}
This problem is also called the Gelfand problem and is a typical example for problems from combustion theory, see for example [12].
Discretization with finite elements
As usual, we first derive the weak formulation for this problem by multiplying with a smooth test function \(v\colon\Omega\to\mathbb{R}\) respecting the boundary condition and integrating over the domain \(\Omega\). Integration by parts and putting the term from the right hand side to the left yields the weak formulation: Find a function \(u\colon\Omega\to\mathbb{R}\) such that for all test functions \(v\) it holds:
\begin{align*}
\int_\Omega \nabla v \cdot \nabla u \,\mathrm{d}x
-
\int_\Omega v \exp(u) \,\mathrm{d}x
=
0.
\end{align*}
Choosing the Lagrangian finite element space \(V_h \dealcoloneq
\bigl\{ v \in C(\overline{\Omega}) : v|_Q \in \mathbb{Q}_p \text{ for all }
Q \in \mathcal{T}_h \bigr\} \cap H_0^1(\Omega)\), which directly incorporates the homogeneous Dirichlet boundary condition, we can define a basis \(\{\varphi_i\}_{i=1,\dots,N}\) and thus it suffices to test only with those basis functions. So the discrete problem reads as follows: Find \(u_h\in V_h\) such that for all \(i=1,\dots,N\) it holds:
\begin{align*}
F(u_h)
\dealcoloneq
\int_\Omega \nabla \varphi_i \cdot \nabla u_h \,\mathrm{d}x
-
\int_\Omega \varphi_i \exp(u_h) \,\mathrm{d}x \stackrel{!}{=} 0.
\end{align*}
As each finite element function is a linear combination of the basis functions \(\{\varphi_i\}_{i=1,\dots,N}\), we can identify the finite element solution by a vector from \(\mathbb{R}^N\) consisting of the unknown values in each degree of freedom (DOF). Thus, we define the nonlinear function \(F\colon\mathbb{R}^N\to\mathbb{R}^N\) representing the discrete nonlinear problem.
To solve this nonlinear problem we use Newton's method. So given an initial guess \(u_h^0\in V_h\), which already fulfills the Dirichlet boundary condition, we determine a sequence of Newton steps \(\bigl( u_h^n \bigr)_n\) by successively applying the following scheme:
\begin{align*}
&\text{Solve for } s_h^n\in V_h: \quad & F'(u_h^n)[s_h^n] &= -F(u_h^n),\\
&\text{Update: } & u_h^{n+1} &= u_h^n + s_h^n.
\end{align*}
So in each Newton step we have to solve a linear problem \(A\,x = b\), where the system matrix \(A\) is represented by the Jacobian \(F'(u_h^n)[\,\cdot\,]\colon\mathbb{R}^N\to\mathbb{R}^N\) and the right hand side \(b\) by the negative residual \(-F(u_h^n)\). The solution vector \(x\) is in that case the Newton update of the \(n\)-th Newton step. Note, that we assume an initial guess \(u_h^0\), which already fulfills the Dirichlet boundary conditions of the problem formulation (in fact this could also be an inhomogeneous Dirichlet boundary condition) and thus the Newton updates \(s_h\) satisfy a homogeneous Dirichlet condition.
Until now we only tested with the basis functions, however, we can also represent any function of \(V_h\) as linear combination of basis functions. More mathematically this means, that every element of \(V_h\) can be identified with a vector \(U\in\mathbb{R}^N\) via the representation formula: \(u_h = \sum_{i=1}^N U_i \varphi_i\). So using this we can give an expression for the discrete Jacobian and the residual:
\begin{align*}
A_{i,j} = \bigl( F'(u_h^n) \bigr)_{i,j}
&=
\int_\Omega \nabla\varphi_i \cdot \nabla \varphi_j \,\mathrm{d} x
-
\int_\Omega \varphi_i \, \exp( u_h ) \varphi_j \,\mathrm{d} x,\\
b_{i} = \bigl( F(u_h^n) \bigr)_{i}
&=
\int_\Omega \nabla\varphi_i \cdot \nabla u_h^n \,\mathrm{d} x
-
\int_\Omega \varphi_i \, \exp( u_h^n ) \,\mathrm{d} x.
\end{align*}
Compared to step-15 we could also have formed the Frech{\'e}t derivative of the nonlinear function corresponding to the strong formulation of the problem and discretized it afterwards. However, in the end we would get the same set of discrete equations.
Numerical linear algebra
Note, how the system matrix, actually the Jacobian, depends on the previous Newton step \(A = F'(u^n)\). Hence we need to tell the function that computes the system matrix about the solution at the last Newton step. In an implementation with a classical assemble_system() function we would gather this information from the last Newton step during assembly by the use of the member functions FEValuesBase::get_function_values() and FEValuesBase::get_function_gradients(). The assemble_system() function would then looks like:
template <int dim>
void GelfandProblem<dim>::assemble_system()
{
system_matrix = 0;
system_rhs = 0;
quadrature_formula,
const unsigned int n_q_points = fe_values.n_quadrature_points;
const unsigned int dofs_per_cell = fe_values.dofs_per_cell;
std::vector<types::global_dof_index> local_dof_indices(dofs_per_cell);
std::vector<Tensor<1, dim>> newton_step_gradients(n_q_points);
std::vector<double> newton_step_values(n_q_points);
for (const auto &cell : dof_handler.active_cell_iterators())
{
cell_rhs = 0.0;
fe_values.reinit(cell);
fe_values.get_function_values(solution, newton_step_values);
fe_values.get_function_gradients(solution, newton_step_gradients);
for (unsigned int q = 0; q < n_q_points; ++q)
{
const double nonlinearity =
std::exp(newton_step_values[q]);
const double dx = fe_values.JxW(q);
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
const double phi_i = fe_values.shape_value(i, q);
for (unsigned int j = 0; j < dofs_per_cell; ++j)
{
const double phi_j = fe_values.shape_value(j, q);
(grad_phi_i * grad_phi_j - phi_i * nonlinearity * phi_j) *
}
cell_rhs(i) += (-grad_phi_i * newton_step_gradients[q] +
phi_i * newton_step_values[q]) *
}
}
cell->get_dof_indices(local_dof_indices);
constraints.distribute_local_to_global(
cell_matrix, cell_rhs, local_dof_indices, system_matrix, system_rhs);
}
}
@ update_values
Shape function values.
@ update_JxW_values
Transformed quadrature weights.
@ update_gradients
Shape function gradients.
void cell_matrix(FullMatrix< double > &M, const FEValuesBase< dim > &fe, const FEValuesBase< dim > &fetest, const ArrayView< const std::vector< double > > &velocity, const double factor=1.)
::VectorizedArray< Number, width > exp(const ::VectorizedArray< Number, width > &)
Since we want to solve this problem without storing a matrix, we need to tell the matrix-free operator this information before we use it. Therefore in the derived class JacobianOperator we will implement a function called evaluate_newton_step, which will process the information of the last Newton step prior to the usage of the matrix-vector implementation. Furthermore we want to use a geometric multigrid (GMG) preconditioner for the linear solver, so in order to apply the multilevel operators we need to pass the last Newton step also to these operators. This is kind of a tricky task, since the vector containing the last Newton step has to be interpolated to all levels of the triangulation. In the code this task will be done by the function MGTransferMatrixFree::interpolate_to_mg(). Note, a fundamental difference to the previous cases, where we set up and used a geometric multigrid preconditioner, is the fact, that we can reuse the MGTransferMatrixFree object for the computation of all Newton steps. So we can save some work here by defining a class variable and using an already set up MGTransferMatrixFree object mg_transfer that was initialized in the setup_system() function.
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::compute_update()
{
solution.update_ghost_values();
system_matrix.evaluate_newton_step(solution);
mg_transfer.interpolate_to_mg(dof_handler, mg_solution, solution);
{
mg_matrices[
level].evaluate_newton_step(mg_solution[
level]);
}
}
const ::parallel::distributed::Triangulation< dim, spacedim > * triangulation
The function evaluating the nonlinearity works basically in the same way as the function evaluate_coefficient from step-37 evaluating a coefficient function. The idea is to use an FEEvaluation object to evaluate the Newton step and store the expression in a table for all cells and all quadrature points:
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::evaluate_newton_step(
{
const unsigned int n_cells = this->data->n_cell_batches();
nonlinear_values.reinit(
n_cells, phi.n_q_points);
for (
unsigned int cell = 0; cell <
n_cells; ++cell)
{
phi.reinit(cell);
phi.read_dof_values_plain(newton_step);
for (unsigned int q = 0; q < phi.n_q_points; ++q)
{
nonlinear_values(cell, q) =
std::exp(phi.get_value(q));
}
}
}
unsigned int n_cells(const internal::TriangulationImplementation::NumberCache< 1 > &c)
As said in step-37 the matrix-free method gets more efficient if we choose a higher order finite element space. Since we want to solve the problem on the \(d\)-dimensional unit ball, it would be good to have an appropriate boundary approximation to overcome convergence issues. For this reason we use an isoparametric approach with the MappingQGeneric class to recover the smooth boundary as well as the mapping for inner cells. In addition, to get a good triangulation in total we make use of the TransfiniteInterpolationManifold.
The commented program
First we include the typical headers of the deal.II library needed for this tutorial:
In particular, we need to include the headers for the matrix-free framework:
And since we want to use a geometric multigrid preconditioner, we need also the multilevel headers:
Finally some common C++ headers for in and output:
#include <fstream>
#include <iostream>
namespace Step66
{
Matrix-free JacobianOperator
In the beginning we define the matrix-free operator for the Jacobian. As a guideline we follow the tutorials step-37 and step-48, where the precise interface of the MatrixFreeOperators::Base class was extensively documented.
Since we want to use the Jacobian as system matrix and pass it to the linear solver as well as to the multilevel preconditioner classes, we derive the JacobianOperator class from the MatrixFreeOperators::Base class, such that we have already the right interface. The two functions we need to override from the base class are the MatrixFreeOperators::Base::apply_add() and the MatrixFreeOperators::Base::compute_diagonal() function. To allow preconditioning with float precision we define the number type as template argument.
As mentioned already in the introduction, we need to evaluate the Jacobian \(F'\) at the last Newton step \(u_h^n\) for the computation of the Newton update \(s_h^n\). To get the information of the last Newton step \(u_h^n\) we do pretty much the same as in step-37, where we stored the values of a coefficient function in a table nonlinear_values once before we use the matrix-free operator. Instead of a function evaluate_coefficient(), we here implement a function evaluate_newton_step().
As additional private member functions of the JacobianOperator we implement the local_apply() and the local_compute_diagonal() function. The first one is the actual worker function for the matrix-vector application, which we pass to the MatrixFree::cell_loop() in the apply_add() function. The later one is the worker function to compute the diagonal, which we pass to the MatrixFreeTools::compute_diagonal() function.
For better readability of the source code we further define an alias for the FEEvaluation object.
template <int dim, int fe_degree, typename number>
class JacobianOperator
Base<dim, LinearAlgebra::distributed::Vector<number>>
{
public:
using value_type = number;
using FECellIntegrator =
JacobianOperator();
virtual void clear() override;
void evaluate_newton_step(
private:
virtual void apply_add(
void
const std::pair<unsigned int, unsigned int> &cell_range) const;
void local_compute_diagonal(FECellIntegrator &integrator) const;
};
The constructor of the JacobianOperator just calls the constructor of the base class MatrixFreeOperators::Base, which is itself derived from the Subscriptor class.
template <int dim, int fe_degree, typename number>
JacobianOperator<dim, fe_degree, number>::JacobianOperator()
{}
The clear() function resets the table holding the values for the nonlinearity and call the clear() function of the base class.
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::clear()
{
nonlinear_values.reinit(0, 0);
clear();
}
Evaluation of the old Newton step
The following evaluate_newton_step() function is based on the evaluate_coefficient() function from step-37. However, it does not evaluate a function object, but evaluates a vector representing a finite element function, namely the last Newton step needed for the Jacobian. Therefore we set up a FEEvaluation object and evaluate the finite element function in the quadrature points with the FEEvaluation::read_dof_values_plain() and FEEvaluation::evaluate() functions. We store the evaluated values of the finite element function directly in the nonlinear_values table.
This will work well and in the local_apply() function we can use the values stored in the table to apply the matrix-vector product. However, we can also optimize the implementation of the Jacobian at this stage. We can directly evaluate the nonlinear function std::exp(newton_step[q]) and store these values in the table. This skips all evaluations of the nonlinearity in each call of the vmult() function.
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::evaluate_newton_step(
{
const unsigned int n_cells = this->data->n_cell_batches();
FECellIntegrator phi(*this->data);
nonlinear_values.reinit(
n_cells, phi.n_q_points);
for (
unsigned int cell = 0; cell <
n_cells; ++cell)
{
phi.reinit(cell);
phi.read_dof_values_plain(newton_step);
for (unsigned int q = 0; q < phi.n_q_points; ++q)
{
nonlinear_values(cell, q) =
std::exp(phi.get_value(q));
}
}
}
Nonlinear matrix-free operator application
Now in the local_apply() function, which actually implements the cell wise action of the system matrix, we can use the information of the last Newton step stored in the table nonlinear_values. The rest of this function is basically the same as in step-37. We set up the FEEvaluation object, gather and evaluate the values and gradients of the input vector src, submit the values and gradients according to the form of the Jacobian and finally call FEEvaluation::integrate_scatter() to perform the cell integration and distribute the local contributions into the global vector dst.
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::local_apply(
const std::pair<unsigned int, unsigned int> & cell_range) const
{
FECellIntegrator phi(data);
for (unsigned int cell = cell_range.first; cell < cell_range.second; ++cell)
{
phi.get_matrix_free().n_cell_batches());
phi.reinit(cell);
phi.gather_evaluate(src,
for (unsigned int q = 0; q < phi.n_q_points; ++q)
{
phi.submit_value(-nonlinear_values(cell, q) * phi.get_value(q), q);
phi.submit_gradient(phi.get_gradient(q), q);
}
dst);
}
}
#define AssertDimension(dim1, dim2)
Next we use MatrixFree::cell_loop() to perform the actual loop over all cells computing the cell contribution to the matrix-vector product.
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::apply_add(
{
this->data->
cell_loop(&JacobianOperator::local_apply,
this, dst, src);
}
void cell_loop(const std::function< void(const MatrixFree< dim, Number, VectorizedArrayType > &, OutVector &, const InVector &, const std::pair< unsigned int, unsigned int > &)> &cell_operation, OutVector &dst, const InVector &src, const bool zero_dst_vector=false) const
Diagonal of the JacobianOperator
The internal worker function local_compute_diagonal() for the computation of the diagonal is similar to the above worker function local_apply(). However, as major difference we do not read values from a input vector or distribute any local results to an output vector. Instead the only input argument is the used FEEvaluation object.
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::local_compute_diagonal(
FECellIntegrator &phi) const
{
phi.get_matrix_free().n_cell_batches());
const unsigned int cell = phi.get_current_cell_index();
for (unsigned int q = 0; q < phi.n_q_points; ++q)
{
phi.submit_value(-nonlinear_values(cell, q) * phi.get_value(q), q);
phi.submit_gradient(phi.get_gradient(q), q);
}
}
Finally we override the MatrixFreeOperators::Base::compute_diagonal() function of the base class of the JacobianOperator. Although the name of the function suggests just the computation of the diagonal, this function does a bit more. Because we only really need the inverse of the matrix diagonal elements for the Chebyshev smoother of the multigrid preconditioner, we compute the diagonal and store the inverse elements. Therefore we first initialize the inverse_diagonal_entries. Then we compute the diagonal by passing the worker function local_compute_diagonal() to the MatrixFreeTools::compute_diagonal() function. In the end we loop over the diagonal and invert the elements by hand. Note, that during this loop we catch the constrained DOFs and set them manually to one.
template <int dim, int fe_degree, typename number>
{
this->inverse_diagonal_entries.reset(
this->inverse_diagonal_entries->get_vector();
inverse_diagonal,
&JacobianOperator::local_compute_diagonal,
this);
for (auto &diagonal_element : inverse_diagonal)
{
diagonal_element = (
std::abs(diagonal_element) > 1.0e-10) ?
(1.0 / diagonal_element) :
1.0;
}
}
void initialize_dof_vector(VectorType &vec, const unsigned int dof_handler_index=0) const
::VectorizedArray< Number, width > abs(const ::VectorizedArray< Number, width > &)
GelfandProblem class
After implementing the matrix-free operators we can now define the solver class for the Gelfand problem. This class is based on the common structure of all previous tutorial programs, in particular it is based on step-15, solving also a nonlinear problem. Since we are using the matrix-free framework, we no longer need an assemble_system function any more, instead the information of the matrix is rebuilt in every call of the vmult() function. However, for the application of the Newton scheme we need to assemble the right hand side of the linearized problems and compute the residuals. Therefore, we implement an additional function evaluate_residual(), which we later call in the assemble_rhs() and the compute_residual() function. Finally, the typical solve() function here implements the Newton method, whereas the solution of the linearized system is computed in the function compute_update(). As the MatrixFree framework handles the polynomial degree of the Lagrangian finite element method as a template parameter, we declare it also as a template parameter for the problem solver class.
template <int dim, int fe_degree>
class GelfandProblem
{
public:
GelfandProblem();
private:
void make_grid();
void setup_system();
void evaluate_residual(
void local_evaluate_residual(
const std::pair<unsigned int, unsigned int> & cell_range) const;
void assemble_rhs();
double compute_residual(const double alpha);
void compute_update();
void solve();
double compute_solution_norm() const;
void output_results(const unsigned int cycle) const;
void run(const Iterator &begin, const typename identity< Iterator >::type &end, Worker worker, Copier copier, const ScratchData &sample_scratch_data, const CopyData &sample_copy_data, const unsigned int queue_length, const unsigned int chunk_size)
For the parallel computation we define a parallel::distributed::Triangulation. As the computational domain is a circle in 2D and a ball in 3D, we assign in addition to the SphericalManifold for boundary cells a TransfiniteInterpolationManifold object for the mapping of the inner cells, which takes care of the inner cells. In this example we use an isoparametric finite element approach and thus use the MappingQGeneric class. Note, that we could also create an instance of the MappingQ class and set the use_mapping_q_on_all_cells flags in the contructor call to true. For further details on the connection of MappingQ and MappingQGeneric you may read the detailed description of these classes.
As usual we then define the Lagrangian finite elements FE_Q and a DoFHandler.
For the linearized discrete system we define an AffineConstraints objects and the system_matrix, which is in this example represented as a matrix-free operator.
using SystemMatrixType = JacobianOperator<dim, fe_degree, double>;
SystemMatrixType system_matrix;
The multilevel object is also based on the matrix-free operator for the Jacobian. Since we need to evaluate the Jacobian with the last Newton step, we also need to evaluate the level operator with the last Newton step for the preconditioner. Thus in addition to mg_matrices, we also need a MGLevelObject to store the interpolated solution vector on each level. As in step-37 we use float precision for the preconditioner. Moreover, we define the MGTransferMatrixFree object as a class variable, since we need to set it up only once when the triangulation has changed and can then use it again in each Newton step.
using LevelMatrixType = JacobianOperator<dim, fe_degree, float>;
Of course we also need vectors holding the solution, the newton_update and the system_rhs. In that way we can always store the last Newton step in the solution vector and just add the update to get the next Newton step.
Finally we have a variable for the number of iterations of the linear solver.
unsigned int linear_iterations;
For the output in programs running in parallel with MPI, we use the ConditionalOStream class to avoid multiple output of the same data by different MPI ranks.
Finally for the time measurement we use a TimerOutput object, which prints the elapsed CPU and wall times for each function in a nicely formatted table after the program has finished.
The constructor of the GelfandProblem initializes the class variables. In particular, we set up the multilevel support for the parallel::distributed::Triangulation, set the mapping degree equal to the finite element degree, initialize the ConditionalOStream and tell the TimerOutput that we want to see the wall times only on demand.
template <int dim, int fe_degree>
GelfandProblem<dim, fe_degree>::GelfandProblem()
, mapping(fe_degree)
, fe(fe_degree)
, computing_timer(MPI_COMM_WORLD,
pcout,
{}
@ construct_multigrid_hierarchy
unsigned int this_mpi_process(const MPI_Comm &mpi_communicator)
GelfandProblem::make_grid
As the computational domain we use the dim-dimensional unit ball. We follow the instructions for the TransfiniteInterpolationManifold class and also assign a SphericalManifold for the boundary. Finally, we refine the initial mesh 3 - dim times globally.
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::make_grid()
{
}
void initialize(const Triangulation< dim, spacedim > &triangulation)
void hyper_ball(Triangulation< dim > &tria, const Point< dim > ¢er=Point< dim >(), const double radius=1., const bool attach_spherical_manifold_on_boundary_cells=false)
GelfandProblem::setup_system
The setup_system() function is quasi identical to the one in step-37. The only differences are obviously the time measurement with only one TimerOutput::Scope instead of measuring each part individually, and more importantly the initialization of the MGLevelObject for the interpolated solution vector of the previous Newton step. Another important change is the setup of the MGTransferMatrixFree object, which we can reuse in each Newton step as the triangulation will not be not changed.
Note how we can use the same MatrixFree object twice, for the JacobianOperator and the multigrid preconditioner.
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::setup_system()
{
system_matrix.clear();
mg_matrices.clear_elements();
dof_handler.distribute_dofs(fe);
dof_handler.distribute_mg_dofs();
constraints.clear();
constraints.reinit(locally_relevant_dofs);
0,
constraints);
constraints.close();
{
auto system_mf_storage = std::make_shared<MatrixFree<dim, double>>();
system_mf_storage->reinit(mapping,
dof_handler,
constraints,
additional_data);
system_matrix.initialize(system_mf_storage);
}
system_matrix.initialize_dof_vector(solution);
system_matrix.initialize_dof_vector(newton_update);
system_matrix.initialize_dof_vector(system_rhs);
mg_matrices.resize(0, nlevels - 1);
mg_solution.resize(0, nlevels - 1);
std::set<types::boundary_id> dirichlet_boundary;
dirichlet_boundary.insert(0);
mg_constrained_dofs.initialize(dof_handler);
mg_constrained_dofs.make_zero_boundary_constraints(dof_handler,
dirichlet_boundary);
mg_transfer.initialize_constraints(mg_constrained_dofs);
mg_transfer.build(dof_handler);
{
relevant_dofs);
level_constraints.
reinit(relevant_dofs);
mg_constrained_dofs.get_boundary_indices(
level));
level_constraints.
close();
auto mg_mf_storage_level = std::make_shared<MatrixFree<dim, float>>();
mg_mf_storage_level->reinit(mapping,
dof_handler,
level_constraints,
additional_data);
mg_matrices[
level].initialize(mg_mf_storage_level,
mg_constrained_dofs,
mg_matrices[
level].initialize_dof_vector(mg_solution[
level]);
}
}
void reinit(const IndexSet &local_constraints=IndexSet())
void add_lines(const std::vector< bool > &lines)
@ update_quadrature_points
Transformed quadrature points.
void make_hanging_node_constraints(const DoFHandler< dim, spacedim > &dof_handler, AffineConstraints< number > &constraints)
TasksParallelScheme tasks_parallel_scheme
UpdateFlags mapping_update_flags
GelfandProblem::evaluate_residual
Next we implement a function which evaluates the nonlinear discrete residual for a given input vector ( \(\texttt{dst} = F(\texttt{src})\)). This function is then used for the assembly of the right hand side of the linearized system and later for the computation of the residual of the next Newton step to check if we already reached the error tolerance. As this function should not affect any class variable we define it as a constant function. Internally we exploit the fast finite element evaluation through the FEEvaluation class and the MatrixFree::cell_loop(), similar to apply_add() function of the JacobianOperator.
First we create a pointer to the MatrixFree object, which is stored in the system_matrix. Then we pass the worker function local_evaluate_residual() for the cell wise evaluation of the residual together with the input and output vector to the MatrixFree::cell_loop(). In addition, we enable the zero out of the output vector in the loop, which is more efficient than calling dst = 0.0 separately before.
Note that with this approach we do not have to take care about the MPI related data exchange, since all the bookkeeping is done by the MatrixFree::cell_loop().
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::evaluate_residual(
{
auto matrix_free = system_matrix.get_matrix_free();
matrix_free->cell_loop(
&GelfandProblem::local_evaluate_residual, this, dst, src, true);
}
GelfandProblem::local_evaluate_residual
This is the internal worker function for the evaluation of the residual. Essentially it has the same structure as the local_apply() function of the JacobianOperator and evaluates the residual for the input vector src on the given set of cells cell_range. The difference to the above mentioned local_apply() function is, that we split the FEEvaluation::gather_evaluate() function into FEEvaluation::read_dof_values_plain() and FEEvaluation::evaluate(), since the input vector might have constrained DOFs.
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::local_evaluate_residual(
const std::pair<unsigned int, unsigned int> & cell_range) const
{
for (unsigned int cell = cell_range.first; cell < cell_range.second; ++cell)
{
phi.reinit(cell);
phi.read_dof_values_plain(src);
for (unsigned int q = 0; q < phi.n_q_points; ++q)
{
phi.submit_value(-
std::exp(phi.get_value(q)), q);
phi.submit_gradient(phi.get_gradient(q), q);
}
dst);
}
}
GelfandProblem::assemble_rhs
Using the above function evaluate_residual() to evaluate the nonlinear residual, the assembly of the right hand side of the linearized system becomes now a very easy task. We just call the evaluate_residual() function and multiply the result with minus one.
Experiences show that using the FEEvaluation class is much faster than a classical implementation with FEValues and co.
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::assemble_rhs()
{
evaluate_residual(system_rhs, solution);
system_rhs *= -1.0;
}
GelfandProblem::compute_residual
According to step-15 the following function computes the norm of the nonlinear residual for the solution \(u_h^n + \alpha s_h^n\) with the help of the evaluate_residual() function. The Newton step length \(\alpha\) becomes important if we would use an adaptive version of the Newton method. Then for example we would compute the residual for different step lengths and compare the residuals. However, for our problem the full Newton step with \(\alpha=1\) is the best we can do. An adaptive version of Newton's method becomes interesting if we have no good initial value. Note that in theory Newton's method converges with quadratic order, but only if we have an appropriate initial value. For unsuitable initial values the Newton method diverges even with quadratic order. A common way is then to use a damped version \(\alpha<1\) until the Newton step is good enough and the full Newton step can be performed. This was also discussed in step-15.
template <int dim, int fe_degree>
double GelfandProblem<dim, fe_degree>::compute_residual(const double alpha)
{
system_matrix.initialize_dof_vector(residual);
system_matrix.initialize_dof_vector(evaluation_point);
evaluation_point = solution;
{
evaluation_point.
add(alpha, newton_update);
}
evaluate_residual(residual, evaluation_point);
}
virtual void add(const Number a) override
virtual real_type l2_norm() const override
SymmetricTensor< 2, dim, Number > e(const Tensor< 2, dim, Number > &F)
GelfandProblem::compute_update
In order to compute the Newton updates in each Newton step we solve the linear system with the CG algorithm together with a geometric multigrid preconditioner. For this we first set up the PreconditionMG object with a Chebyshev smoother like we did in step-37.
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::compute_update()
{
We remember that the Jacobian depends on the last Newton step stored in the solution vector. So we update the ghost values of the Newton step and pass it to the JacobianOperator to store the information.
solution.update_ghost_values();
system_matrix.evaluate_newton_step(solution);
Next we also have to pass the last Newton step to the multilevel operators. Therefore, we need to interpolate the Newton step to all levels of the triangulation. This is done with the MGTransferMatrixFree::interpolate_to_mg().
mg_transfer.interpolate_to_mg(dof_handler, mg_solution, solution);
Now we can set up the preconditioner. We define the smoother and pass the interpolated vectors of the Newton step to the multilevel operators.
using SmootherType =
mg_smoother;
{
{
smoother_data[
level].smoothing_range = 15.;
smoother_data[
level].degree = 4;
smoother_data[
level].eig_cg_n_iterations = 10;
}
else
{
smoother_data[0].smoothing_range = 1
e-3;
smoother_data[0].eig_cg_n_iterations = mg_matrices[0].m();
}
mg_matrices[
level].evaluate_newton_step(mg_solution[
level]);
mg_matrices[
level].compute_diagonal();
smoother_data[
level].preconditioner =
mg_matrices[
level].get_matrix_diagonal_inverse();
}
mg_smoother.initialize(mg_matrices, smoother_data);
mg_coarse;
mg_matrices);
mg_interface_matrices;
{
mg_interface_matrices[
level].initialize(mg_matrices[
level]);
}
mg_interface_matrices);
mg_matrix, mg_coarse, mg_transfer, mg_smoother, mg_smoother);
mg.set_edge_matrices(mg_interface, mg_interface);
preconditioner(dof_handler,
mg, mg_transfer);
void initialize(const MGSmootherBase< VectorType > &coarse_smooth)
void resize(const unsigned int new_minlevel, const unsigned int new_maxlevel, Args &&... args)
static const unsigned int invalid_unsigned_int
Finally we set up the SolverControl and the SolverCG to solve the linearized problem for the current Newton update. An important fact of the implementation of SolverCG or also SolverGMRES is, that the vector holding the solution of the linear system (here newton_update) can be used to pass a starting value. In order to start the iterative solver always with a zero vector we reset the newton_update explicitly before calling SolverCG::solve(). Afterwards we distribute the Dirichlet boundary conditions stored in constraints and store the number of iteration steps for the later output.
newton_update = 0.0;
cg.solve(system_matrix, newton_update, system_rhs, preconditioner);
constraints.distribute(newton_update);
linear_iterations = solver_control.last_step();
Then for bookkeeping we zero out the ghost values.
solution.zero_out_ghost_values();
}
GelfandProblem::solve
Now we implement the actual Newton solver for the nonlinear problem.
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::solve()
{
We define a maximal number of Newton steps and tolerances for the convergence criterion. Usually, with good starting values, the Newton method converges in three to six steps, so maximal ten steps should be totally sufficient. As tolerances we use \(\|F(u^n_h)\|<\text{TOL}_f =
10^{-12}\) for the norm of the residual and \(\|s_h^n\| < \text{TOL}_x =
10^{-10}\) for the norm of the Newton update. This seems a bit over the top, but we will see that, for our example, we will achieve these tolerances after a few steps.
const unsigned int itmax = 10;
const double TOLf = 1
e-12;
const double TOLx = 1
e-10;
Now we start the actual Newton iteration.
for (unsigned int newton_step = 1; newton_step <= itmax; ++newton_step)
{
We assemble the right hand side of the linearized problem and compute the Newton update.
assemble_rhs();
compute_update();
Then we compute the errors, namely the norm of the Newton update and the residual. Note that at this point one could incorporate a step size control for the Newton method by varying the input parameter \(\alpha\) for the compute_residual function. However, here we just use \(\alpha\) equal to one for a plain Newton iteration.
const double ERRx = newton_update.l2_norm();
const double ERRf = compute_residual(1.0);
Next we advance the Newton step by adding the Newton update to the current Newton step.
solution.add(1.0, newton_update);
A short output will inform us on the current Newton step.
pcout << " Nstep " << newton_step << ", errf = " << ERRf
<< ", errx = " << ERRx << ", it = " << linear_iterations
<< std::endl;
After each Newton step we check the convergence criteria. If at least one of those is fulfilled we are done and end the loop. If we haven't found a satisfying solution after the maximal amount of Newton iterations, we inform the user about this shortcoming.
if (ERRf < TOLf || ERRx < TOLx)
{
pcout << "Convergence step " << newton_step << " value " << ERRf
<<
" (used wall time: " << solver_timer.
wall_time() <<
" s)" << std::endl;
break;
}
else if (newton_step == itmax)
{
pcout << "WARNING: No convergence of Newton's method after "
<< newton_step << " steps." << std::endl;
break;
}
}
}
GelfandProblem::compute_solution_norm
The computation of the H1-seminorm of the solution can be done in the same way as in step-59. We update the ghost values and use the function VectorTools::integrate_difference(). In the end we gather all computations from all MPI ranks and return the norm.
template <int dim, int fe_degree>
double GelfandProblem<dim, fe_degree>::compute_solution_norm() const
{
solution.update_ghost_values();
dof_handler,
solution,
norm_per_cell,
solution.zero_out_ghost_values();
norm_per_cell,
}
GelfandProblem::output_results
We generate the graphical output files in vtu format together with a pvtu master file at once by calling the DataOut::write_vtu_with_pvtu_record() function in the same way as in step-37. In addition, as in step-40, we query the types::subdomain_id of each cell and write the distribution of the triangulation among the MPI ranks into the output file. Finally, we generate the patches of the solution by calling DataOut::build_patches(). However, since we have a computational domain with a curved boundary, we additionally pass the mapping and the finite element degree as number of subdivision. But this is still not enough for the correct representation of the solution, for example in ParaView, because we attached a TransfiniteInterpolationManifold to the inner cells, which results in curved cells in the interior. Therefore we pass as third argument the DataOut::curved_inner_cells option, such that also the inner cells use the corresponding manifold description to build the patches.
Note that we could handle the higher order elements with the flag DataOutBase::VtkFlags::write_higher_order_cells. However, due to the limited compatibility to previous version of ParaView and the missing support by VisIt, we left this option for a future version.
template <int dim, int fe_degree>
void
GelfandProblem<dim, fe_degree>::output_results(const unsigned int cycle) const
{
return;
solution.update_ghost_values();
for (unsigned int i = 0; i < subdomain.size(); ++i)
{
}
fe.degree,
"./",
"solution_" +
std::to_string(dim) +
"d", cycle, MPI_COMM_WORLD, 3);
solution.zero_out_ghost_values();
}
void attach_dof_handler(const DoFHandlerType &)
void add_data_vector(const VectorType &data, const std::vector< std::string > &names, const DataVectorType type=type_automatic, const std::vector< DataComponentInterpretation::DataComponentInterpretation > &data_component_interpretation=std::vector< DataComponentInterpretation::DataComponentInterpretation >())
virtual void build_patches(const unsigned int n_subdivisions=0)
std::string write_vtu_with_pvtu_record(const std::string &directory, const std::string &filename_without_extension, const unsigned int counter, const MPI_Comm &mpi_communicator, const unsigned int n_digits_for_counter=numbers::invalid_unsigned_int, const unsigned int n_groups=0) const
ZlibCompressionLevel compression_level
std::string to_string(const T &t)
void set_flags(const FlagType &flags)
GelfandProblem::run
The last missing function of the solver class for the Gelfand problem is the run function. In the beginning we print information about the system specifications and the finite element space we use. The problem is solved several times on a successively refined mesh.
template <int dim, int fe_degree>
{
{
const unsigned int n_ranks =
const unsigned int n_vect_bits = 8 * sizeof(double) * n_vect_doubles;
" MPI process" + (n_ranks > 1 ? "es" : "");
std::string VEC_header =
"), VECTORIZATION_LEVEL=" +
std::string SOL_header = "Finite element space: " + fe.get_name();
pcout << std::string(80, '=') << std::endl;
pcout << DAT_header << std::endl;
pcout << std::string(80, '-') << std::endl;
pcout << MPI_header << std::endl;
pcout << VEC_header << std::endl;
pcout << SOL_header << std::endl;
pcout << std::string(80, '=') << std::endl;
}
for (unsigned int cycle = 0; cycle < 9 - dim; ++cycle)
{
pcout << std::string(80, '-') << std::endl;
pcout << "Cycle " << cycle << std::endl;
pcout << std::string(80, '-') << std::endl;
static constexpr std::size_t size()
#define DEAL_II_COMPILER_VECTORIZATION_LEVEL
unsigned int n_mpi_processes(const MPI_Comm &mpi_communicator)
const std::string get_current_vectorization_level()
The first task in actually solving the problem is to generate or refine the triangulation.
if (cycle == 0)
{
make_grid();
}
else
{
}
Now we set up the system and solve the problem. These steps are accompanied by time measurement and textual output.
pcout << "Set up system..." << std::endl;
setup_system();
pcout <<
" Triangulation: " <<
triangulation.n_global_active_cells()
<< " cells" << std::endl;
pcout << " DoFHandler: " << dof_handler.n_dofs() << " DoFs"
<< std::endl;
pcout << std::endl;
pcout << "Solve using Newton's method..." << std::endl;
solve();
pcout << std::endl;
pcout <<
"Time for setup+solve (CPU/Wall) " << timer.
cpu_time() <<
"/"pcout << std::endl;
After the problem was solved we compute the norm of the solution and generate the graphical output files.
pcout << "Output results..." << std::endl;
const double norm = compute_solution_norm();
output_results(cycle);
pcout <<
" H1 seminorm: " <<
norm << std::endl;
pcout << std::endl;
double norm(const FEValuesBase< dim > &fe, const ArrayView< const std::vector< Tensor< 1, dim > > > &Du)
Finally after each cycle we print the timing information.
computing_timer.print_summary();
computing_timer.reset();
}
}
}
The main function
As typical for programs running in parallel with MPI we set up the MPI framework and disable shared-memory parallelization by limiting the number of threads to one. Finally to run the solver for the Gelfand problem we create an object of the GelfandProblem class and call the run function. Exemplarily we solve the problem once in 2D and once in 3D each with fourth-order Lagrangian finite elements.
int main(int argc, char *argv[])
{
try
{
using namespace Step66;
{
GelfandProblem<2, 4> gelfand_problem;
gelfand_problem.run();
}
{
GelfandProblem<3, 4> gelfand_problem;
gelfand_problem.run();
}
}
catch (std::exception &exc)
{
std::cerr << std::endl
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Exception on processing: " << std::endl
<< exc.what() << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
catch (...)
{
std::cerr << std::endl
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Unknown exception!" << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
return 0;
}
Results
The aim of this tutorial step was to demonstrate the solution of a nonlinear PDE with the matrix-free framework.
Program output
Running the program on two processes in release mode via
cmake . && make release && make && mpirun -n 2 ./step-66
gives the following output on the console
================================================================================
START DATE: 2021/5/18, TIME: 16:25:48
--------------------------------------------------------------------------------
Running with 2 MPI processes
Vectorization over 4 doubles = 256 bits (AVX), VECTORIZATION_LEVEL=2
================================================================================
--------------------------------------------------------------------------------
Cycle 0
--------------------------------------------------------------------------------
Set up system...
Solve using Newton's method...
Nstep 1, errf = 0.00380835, errx = 3.61904, it = 7
Nstep 2, errf = 3.80167e-06, errx = 0.104353, it = 6
Nstep 3, errf = 3.97939e-12, errx = 0.00010511, it = 4
Nstep 4, errf = 2.28859e-13, errx = 1.07726e-10, it = 1
Convergence step 4 value 2.28859e-13 (used wall time: 0.0096409 s)
Time for setup+solve (CPU/Wall) 0.015617/0.0156447 s
Output results...
H1 seminorm: 0.773426
+---------------------------------------------+------------+------------+
| Total wallclock time elapsed since start | 0.0286s | |
| | | |
| Section | no. calls | wall time | % of total |
+---------------------------------+-----------+------------+------------+
| assemble right hand side | 4 | 9.71e-05s | 0.34% |
| compute residual | 4 | 0.000137s | 0.48% |
| compute update | 4 | 0.00901s | 32% |
| make grid | 1 | 0.00954s | 33% |
| setup system | 1 | 0.00585s | 20% |
| solve | 1 | 0.00966s | 34% |
+---------------------------------+-----------+------------+------------+
.
.
.
--------------------------------------------------------------------------------
Cycle 6
--------------------------------------------------------------------------------
Set up system...
Triangulation: 81920 cells
DoFHandler: 1311745 DoFs
Solve using Newton's method...
Nstep 1, errf = 5.90478e-05, errx = 231.427, it = 9
Nstep 2, errf = 5.89991e-08, errx = 6.67102, it = 6
Nstep 3, errf = 4.28813e-13, errx = 0.0067188, it = 4
Convergence step 3 value 4.28813e-13 (used wall time: 4.82953 s)
Time for setup+solve (CPU/Wall) 6.25094/6.37174 s
Output results...
H1 seminorm: 0.773426
+---------------------------------------------+------------+------------+
| Total wallclock time elapsed since start | 9.04s | |
| | | |
| Section | no. calls | wall time | % of total |
+---------------------------------+-----------+------------+------------+
|
assemble right hand side | 3 | 0.0827s | 0.91% |
| compute residual | 3 | 0.0909s | 1% |
| compute update | 3 | 4.65s | 51% |
| setup system | 1 | 1.54s | 17% |
| solve | 1 | 4.83s | 53% |
+---------------------------------+-----------+------------+------------+
================================================================================
START DATE: 2021/5/18, TIME: 16:26:00
--------------------------------------------------------------------------------
Running with 2 MPI processes
Vectorization over 4 doubles = 256 bits (AVX), VECTORIZATION_LEVEL=2
================================================================================
.
.
.
--------------------------------------------------------------------------------
Cycle 5
--------------------------------------------------------------------------------
Set up system...
Solve using Newton's method...
Nstep 1, errf = 6.30096e-06, errx = 481.74, it = 8
Nstep 2, errf = 4.25607e-10, errx = 4.14315, it = 6
Nstep 3, errf = 7.29563e-13, errx = 0.000321775, it = 2
Convergence step 3 value 7.29563e-13 (used wall time: 133.793 s)
Time for setup+solve (CPU/Wall) 226.809/232.615 s
Output results...
H1 seminorm: 0.588667
+---------------------------------------------+------------+------------+
| Total wallclock time elapsed since start | 390s | |
| | | |
| Section | no. calls | wall time | % of total |
+---------------------------------+-----------+------------+------------+
| assemble right hand side | 3 | 2.06s | 0.53% |
| compute residual | 3 | 2.46s | 0.63% |
| compute update | 3 | 129s | 33% |
| setup system | 1 | 98.8s | 25% |
| solve | 1 | 134s | 34% |
+---------------------------------+-----------+------------+------------+
void assemble(const MeshWorker::DoFInfoBox< dim, DOFINFO > &dinfo, A *assembler)
TriangulationBase< dim, spacedim > Triangulation
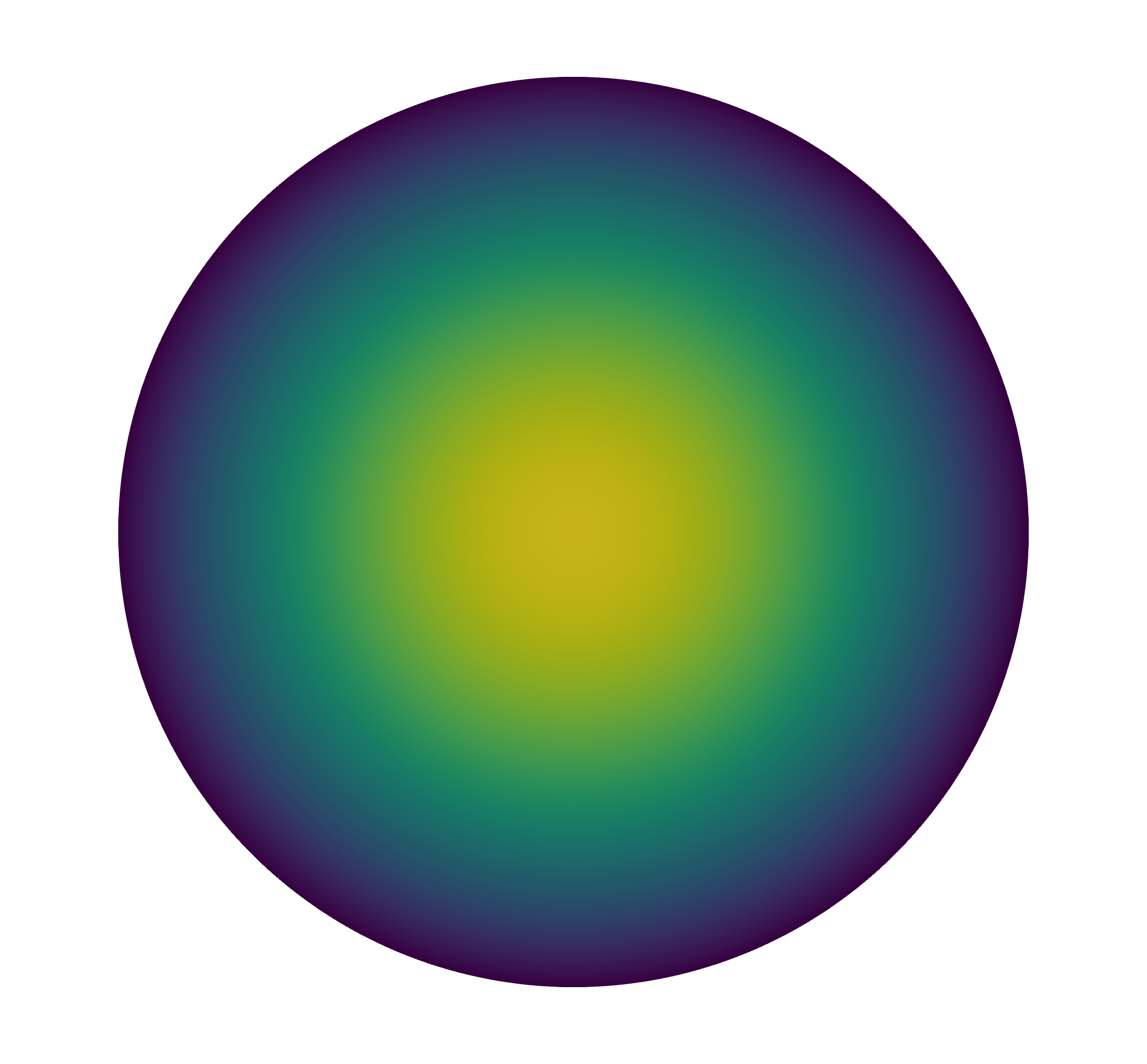
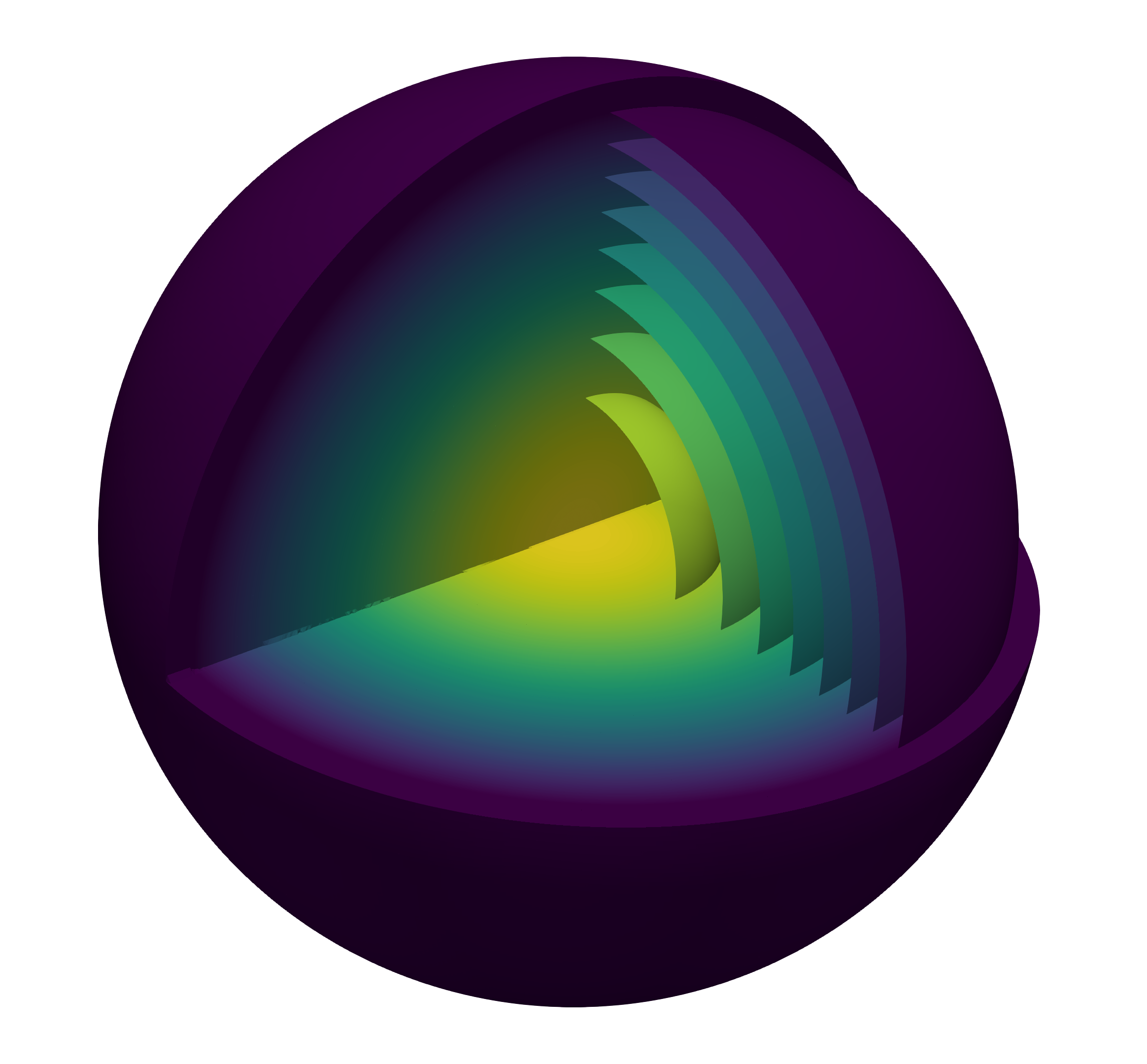
We show the solution for the two- and three-dimensional problem in the following figure.
Newton solver
In the program output above we find some interesting information about the Newton iterations. The terminal output in each refinement cycle presents detailed diagnostics of the Newton method, which show first of all the number of Newton steps and for each step the norm of the residual \(\|F(u_h^{n+1})\|\), the norm of the Newton update \(\|s_h^n\|\), and the number of CG iterations it.
We observe that for all cases the Newton method converges in approximately three to four steps, which shows the quadratic convergence of the Newton method with a full step length \(\alpha = 1\). However, be aware that for a badly chosen initial guess \(u_h^0\), the Newton method will also diverge quadratically. Usually if you do not have an appropriate initial guess, you try a few damped Newton steps with a reduced step length \(\alpha < 1\) until the Newton step is again in the quadratic convergence domain. This damping and relaxation of the Newton step length truly requires a more sophisticated implementation of the Newton method, which we designate to you as a possible extension of the tutorial.
Furthermore, we see that the number of CG iterations is approximately constant with successive mesh refinements and an increasing number of DoFs. This is of course due to the geometric multigrid preconditioner and similar to the observations made in other tutorials that use this method, e.g., step-16 and step-37. Just to give an example, in the three-dimensional case after five refinements, we have approximately 14.7 million distributed DoFs with fourth-order Lagrangian finite elements, but the number of CG iterations is still less than ten.
In addition, there is one more very useful optimization that we applied and that should be mentioned here. In the compute_update() function we explicitly reset the vector holding the Newton update before passing it as the output vector to the solver. In that case we use a starting value of zero for the CG method, which is more suitable than the previous Newton update, the actual content of the newton_update before resetting, and thus reduces the number of CG iterations by a few steps.
Possibilities for extensions
A couple of possible extensions are available concerning minor updates fo the present code as well as a deeper numerical investigation of the Gelfand problem.
More sophisticated Newton iteration
Beside a step size controlled version of the Newton iteration as mentioned already in step-15, one could also implement a more flexible stopping criterion for the Newton iteration. For example one could replace the fixed tolerances for the residual TOLf and for the Newton updated TOLx and implement a mixed error control with a given absolute and relative tolerance, such that the Newton iteration exists with success as, e.g.,
\begin{align*}
\|F(u_h^{n+1})\| \leq \texttt{RelTol} \|u_h^{n+1}\| + \texttt{AbsTol}.
\end{align*}
For more advanced applications with many nonlinear systems to solve, for example at each time step for a time-dependent problem, it turns out that it is not necessary to set up and assemble the Jacobian anew at every single Newton step or even for each time step. Instead, the existing Jacobian from a previous step can be used for the Newton iteration. The Jacobian is then only rebuilt if, for example, the Newton iteration converges too slowly. Such an idea yields a quasi-Newton method. Admittedly, when using the matrix-free framework, the assembly of the Jacobian is omitted anyway, but with in this way one can try to optimize the reassembly of the geometric multigrid preconditioner. Remember that each time the solution from the old Newton step must be distributed to all levels and the mutligrid preconditioner must be reinitialized.
Parallel scalability and thread parallelism
In the results section of step-37 and others, the parallel scalability of the matrix-free framework on a large number of processors has already been demonstrated very impressively. In the nonlinear case we consider here, we note that one of the bottlenecks could become the transfer and evaluation of the matrix-free Jacobi operator and its multistage operators in the previous Newton step, since we need to transfer the old solution at all stages in each step. A first parallel scalability analysis in [30] shows quite good strong scalability when the problem size is large enough. However, a more detailed analysis needs to be performed for reliable results. Moreover, the problem has been solved only with MPI so far, without using the possibilities of shared memory parallelization with threads. Therefore, for this example, you could try hybrid parallelization with MPI and threads, such as described in step-48.
Comparison to matrix-based methods
Analogously to step-50 and the mentioned possible extension of step-75, you can convince yourself which method is faster.
Eigenvalue problem
One can consider the corresponding eigenvalue problem, which is called Bratu problem. For example, if we define a fixed eigenvalue \(\lambda\in[0,6]\), we can compute the corresponding discrete eigenfunction. You will notice that the number of Newton steps will increase with increasing \(\lambda\). To reduce the number of Newton steps you can use the following trick: start from a certain \(\lambda\), compute the eigenfunction, increase \(\lambda=\lambda +
\delta_\lambda\), and then use the previous solution as an initial guess for the Newton iteration. In the end you can plot the \(H^1(\Omega)\)-norm over the eigenvalue \(\lambda \mapsto \|u_h\|_{H^1(\Omega)}\). What do you observe for further increasing \(\lambda>7\)?
The plain program
#include <fstream>
#include <iostream>
namespace Step66
{
template <int dim, int fe_degree, typename number>
class JacobianOperator
Base<dim, LinearAlgebra::distributed::Vector<number>>
{
public:
using value_type = number;
using FECellIntegrator =
JacobianOperator();
virtual void clear() override;
void evaluate_newton_step(
private:
virtual void apply_add(
void
const std::pair<unsigned int, unsigned int> &cell_range) const;
void local_compute_diagonal(FECellIntegrator &integrator) const;
};
template <int dim, int fe_degree, typename number>
JacobianOperator<dim, fe_degree, number>::JacobianOperator()
{}
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::clear()
{
nonlinear_values.
reinit(0, 0);
clear();
}
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::evaluate_newton_step(
{
const unsigned int n_cells = this->data->n_cell_batches();
FECellIntegrator phi(*this->data);
nonlinear_values.reinit(
n_cells, phi.n_q_points);
for (
unsigned int cell = 0; cell <
n_cells; ++cell)
{
phi.reinit(cell);
phi.read_dof_values_plain(newton_step);
for (unsigned int q = 0; q < phi.n_q_points; ++q)
{
nonlinear_values(cell, q) =
std::exp(phi.get_value(q));
}
}
}
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::local_apply(
const std::pair<unsigned int, unsigned int> & cell_range) const
{
FECellIntegrator phi(data);
for (unsigned int cell = cell_range.first; cell < cell_range.second; ++cell)
{
phi.get_matrix_free().n_cell_batches());
phi.reinit(cell);
phi.gather_evaluate(src,
for (unsigned int q = 0; q < phi.n_q_points; ++q)
{
phi.submit_value(-nonlinear_values(cell, q) * phi.get_value(q), q);
phi.submit_gradient(phi.get_gradient(q), q);
}
dst);
}
}
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::apply_add(
{
this->data->
cell_loop(&JacobianOperator::local_apply,
this, dst, src);
}
template <int dim, int fe_degree, typename number>
void JacobianOperator<dim, fe_degree, number>::local_compute_diagonal(
FECellIntegrator &phi) const
{
phi.get_matrix_free().n_cell_batches());
const unsigned int cell = phi.get_current_cell_index();
for (unsigned int q = 0; q < phi.n_q_points; ++q)
{
phi.submit_value(-nonlinear_values(cell, q) * phi.get_value(q), q);
phi.submit_gradient(phi.get_gradient(q), q);
}
}
template <int dim, int fe_degree, typename number>
{
this->inverse_diagonal_entries.reset(
this->inverse_diagonal_entries->get_vector();
inverse_diagonal,
&JacobianOperator::local_compute_diagonal,
this);
for (auto &diagonal_element : inverse_diagonal)
{
diagonal_element = (
std::abs(diagonal_element) > 1.0e-10) ?
(1.0 / diagonal_element) :
1.0;
}
}
template <int dim, int fe_degree>
class GelfandProblem
{
public:
GelfandProblem();
private:
void make_grid();
void setup_system();
void evaluate_residual(
void local_evaluate_residual(
const std::pair<unsigned int, unsigned int> & cell_range) const;
void assemble_rhs();
double compute_residual(const double alpha);
void compute_update();
void solve();
double compute_solution_norm() const;
void output_results(const unsigned int cycle) const;
using SystemMatrixType = JacobianOperator<dim, fe_degree, double>;
SystemMatrixType system_matrix;
using LevelMatrixType = JacobianOperator<dim, fe_degree, float>;
unsigned int linear_iterations;
};
template <int dim, int fe_degree>
GelfandProblem<dim, fe_degree>::GelfandProblem()
, mapping(fe_degree)
, fe(fe_degree)
, computing_timer(MPI_COMM_WORLD,
pcout,
{}
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::make_grid()
{
}
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::setup_system()
{
system_matrix.clear();
mg_matrices.clear_elements();
dof_handler.distribute_dofs(fe);
dof_handler.distribute_mg_dofs();
constraints.clear();
constraints.reinit(locally_relevant_dofs);
0,
constraints);
constraints.close();
{
auto system_mf_storage = std::make_shared<MatrixFree<dim, double>>();
system_mf_storage->reinit(mapping,
dof_handler,
constraints,
additional_data);
system_matrix.initialize(system_mf_storage);
}
system_matrix.initialize_dof_vector(solution);
system_matrix.initialize_dof_vector(newton_update);
system_matrix.initialize_dof_vector(system_rhs);
mg_matrices.resize(0, nlevels - 1);
mg_solution.resize(0, nlevels - 1);
std::set<types::boundary_id> dirichlet_boundary;
dirichlet_boundary.insert(0);
mg_constrained_dofs.initialize(dof_handler);
mg_constrained_dofs.make_zero_boundary_constraints(dof_handler,
dirichlet_boundary);
mg_transfer.initialize_constraints(mg_constrained_dofs);
mg_transfer.build(dof_handler);
{
relevant_dofs);
level_constraints.
reinit(relevant_dofs);
mg_constrained_dofs.get_boundary_indices(
level));
level_constraints.
close();
auto mg_mf_storage_level = std::make_shared<MatrixFree<dim, float>>();
mg_mf_storage_level->reinit(mapping,
dof_handler,
level_constraints,
additional_data);
mg_matrices[
level].initialize(mg_mf_storage_level,
mg_constrained_dofs,
mg_matrices[
level].initialize_dof_vector(mg_solution[
level]);
}
}
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::evaluate_residual(
{
auto matrix_free = system_matrix.get_matrix_free();
matrix_free->cell_loop(
&GelfandProblem::local_evaluate_residual, this, dst, src, true);
}
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::local_evaluate_residual(
const std::pair<unsigned int, unsigned int> & cell_range) const
{
for (unsigned int cell = cell_range.first; cell < cell_range.second; ++cell)
{
phi.reinit(cell);
phi.read_dof_values_plain(src);
for (unsigned int q = 0; q < phi.n_q_points; ++q)
{
phi.submit_value(-
std::exp(phi.get_value(q)), q);
phi.submit_gradient(phi.get_gradient(q), q);
}
dst);
}
}
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::assemble_rhs()
{
evaluate_residual(system_rhs, solution);
system_rhs *= -1.0;
}
template <int dim, int fe_degree>
double GelfandProblem<dim, fe_degree>::compute_residual(const double alpha)
{
system_matrix.initialize_dof_vector(residual);
system_matrix.initialize_dof_vector(evaluation_point);
evaluation_point = solution;
{
evaluation_point.
add(alpha, newton_update);
}
evaluate_residual(residual, evaluation_point);
}
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::compute_update()
{
solution.update_ghost_values();
system_matrix.evaluate_newton_step(solution);
mg_transfer.interpolate_to_mg(dof_handler, mg_solution, solution);
using SmootherType =
mg_smoother;
{
{
smoother_data[
level].smoothing_range = 15.;
smoother_data[
level].degree = 4;
smoother_data[
level].eig_cg_n_iterations = 10;
}
else
{
smoother_data[0].smoothing_range = 1
e-3;
smoother_data[0].eig_cg_n_iterations = mg_matrices[0].m();
}
mg_matrices[
level].evaluate_newton_step(mg_solution[
level]);
mg_matrices[
level].compute_diagonal();
smoother_data[
level].preconditioner =
mg_matrices[
level].get_matrix_diagonal_inverse();
}
mg_smoother.initialize(mg_matrices, smoother_data);
mg_coarse;
mg_matrices);
mg_interface_matrices;
{
mg_interface_matrices[
level].initialize(mg_matrices[
level]);
}
mg_interface_matrices);
mg_matrix, mg_coarse, mg_transfer, mg_smoother, mg_smoother);
mg.set_edge_matrices(mg_interface, mg_interface);
preconditioner(dof_handler,
mg, mg_transfer);
newton_update = 0.0;
cg.solve(system_matrix, newton_update, system_rhs, preconditioner);
constraints.distribute(newton_update);
linear_iterations = solver_control.last_step();
solution.zero_out_ghost_values();
}
template <int dim, int fe_degree>
void GelfandProblem<dim, fe_degree>::solve()
{
const unsigned int itmax = 10;
const double TOLf = 1
e-12;
const double TOLx = 1
e-10;
for (unsigned int newton_step = 1; newton_step <= itmax; ++newton_step)
{
assemble_rhs();
compute_update();
const double ERRx = newton_update.l2_norm();
const double ERRf = compute_residual(1.0);
solution.add(1.0, newton_update);
pcout << " Nstep " << newton_step << ", errf = " << ERRf
<< ", errx = " << ERRx << ", it = " << linear_iterations
<< std::endl;
if (ERRf < TOLf || ERRx < TOLx)
{
pcout << "Convergence step " << newton_step << " value " << ERRf
<<
" (used wall time: " << solver_timer.
wall_time() <<
" s)" << std::endl;
break;
}
else if (newton_step == itmax)
{
pcout << "WARNING: No convergence of Newton's method after "
<< newton_step << " steps." << std::endl;
break;
}
}
}
template <int dim, int fe_degree>
double GelfandProblem<dim, fe_degree>::compute_solution_norm() const
{
dof_handler,
solution,
norm_per_cell,
solution.zero_out_ghost_values();
norm_per_cell,
}
template <int dim, int fe_degree>
void
GelfandProblem<dim, fe_degree>::output_results(const unsigned int cycle) const
{
return;
solution.update_ghost_values();
for (unsigned int i = 0; i < subdomain.size(); ++i)
{
}
fe.degree,
"./",
"solution_" +
std::to_string(dim) +
"d", cycle, MPI_COMM_WORLD, 3);
solution.zero_out_ghost_values();
}
template <int dim, int fe_degree>
{
{
const unsigned int n_ranks =
const unsigned int n_vect_bits = 8 * sizeof(double) * n_vect_doubles;
" MPI process" + (n_ranks > 1 ? "es" : "");
std::string VEC_header =
"), VECTORIZATION_LEVEL=" +
std::string SOL_header = "Finite element space: " + fe.get_name();
pcout << std::string(80, '=') << std::endl;
pcout << DAT_header << std::endl;
pcout << std::string(80, '-') << std::endl;
pcout << MPI_header << std::endl;
pcout << VEC_header << std::endl;
pcout << SOL_header << std::endl;
pcout << std::string(80, '=') << std::endl;
}
for (unsigned int cycle = 0; cycle < 9 - dim; ++cycle)
{
pcout << std::string(80, '-') << std::endl;
pcout << "Cycle " << cycle << std::endl;
pcout << std::string(80, '-') << std::endl;
if (cycle == 0)
{
make_grid();
}
else
{
}
pcout << "Set up system..." << std::endl;
setup_system();
pcout <<
" Triangulation: " <<
triangulation.n_global_active_cells()
<< " cells" << std::endl;
pcout << " DoFHandler: " << dof_handler.n_dofs() << " DoFs"
<< std::endl;
pcout << std::endl;
pcout << "Solve using Newton's method..." << std::endl;
solve();
pcout << std::endl;
pcout <<
"Time for setup+solve (CPU/Wall) " << timer.
cpu_time() <<
"/" pcout << std::endl;
pcout << "Output results..." << std::endl;
const double norm = compute_solution_norm();
output_results(cycle);
pcout <<
" H1 seminorm: " <<
norm << std::endl;
pcout << std::endl;
computing_timer.print_summary();
computing_timer.reset();
}
}
}
int main(int argc, char *argv[])
{
try
{
using namespace Step66;
{
GelfandProblem<2, 4> gelfand_problem;
gelfand_problem.run();
}
{
GelfandProblem<3, 4> gelfand_problem;
gelfand_problem.run();
}
}
catch (std::exception &exc)
{
std::cerr << std::endl
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Exception on processing: " << std::endl
<< exc.what() << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
catch (...)
{
std::cerr << std::endl
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Unknown exception!" << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
return 0;
}
void update_ghost_values() const
virtual void reinit(const size_type N, const bool omit_zeroing_entries=false)