This program was contributed by Ester Comellas <ester.comellas@upc.edu>.
It comes without any warranty or support by its authors or the authors of deal.II.
This program is part of the deal.II code gallery and consists of the following files (click to inspect):
Pictures from this code gallery program
Annotated version of readme.md
Readme file for nonlinear-poro-viscoelasticity
Overview
We implemented a nonlinear poro-viscoelastic formulation with the aim of characterising brain tissue response to cyclic loading. Our model captures both experimentally observed fluid flow and conditioning aspects of brain tissue behavior in addition to its well-established nonlinear, preconditioning, hysteretic, and tension-compression asymmetric characteristics.
The tissue is modelled as a biphasic material consisting of an immiscible aggregate of a nonlinear viscoelastic solid skeleton saturated with pore fluid. The governing equations are linearised using automatic differentiation and solved monolithically for the unknown solid displacements and fluid pore pressure values.
A detailed description of the formulation, its verification, and the results obtained can be found in:
-
E. Comellas, S. Budday, J.-P. Pelteret, G. A. Holzapfel and P. Steinmann (2020), Modeling the porous and viscous responses of human brain tissue behavior, Computer Methods in Applied Mechanics and Engineering, 113128. DOI: 10.1016/j.cma.2020.113128.
In this paper we show that nonlinear poroelasticity alone can reproduce consolidation experiments, yet it is insufficient to capture stress conditioning due to cyclic loading. We also discuss how the poroelastic response exhibits preconditioning and hysteresis in the fluid flow space, with porous and viscous effects being highly interrelated.
Quick facts about the code
-
Biphasic material following the Theory of Porous Media
-
Nonlinear finite viscoelasticity built on Ogden hyperelasticity
-
Darcy-like fluid flow
-
Spatial discretisation with continuous Q2-P1 Lagrangian finite elements
-
Temporal discretisation with a stable implicit one-step backward differentiation method
-
Newton-Raphson scheme to solve the nonlinear system of governing equations
-
Forward mode automatic differentiation with the number of derivative components chosen at run-time (Sacado library within Trilinos package) to linearise the governing equations (and, implicitly, the constitutive laws)
-
Trilinos direct solver for the (non-symmetric) linear system of equations using a monolithic scheme
-
Parallelization through Threading Building Blocks and across nodes via MPI (using Trilinos linear algebra)
-
Based on step-44 and the code gallery contributions 'Quasi-Static Finite-Strain Compressible Elasticity' and 'Quasi-Static Finite-Strain Quasi-incompressible Visco-elasticity'
-
Only works in 3D
Running the code
Requirements
-
MPI and Trilinos (built with the Sacado library) must be enabled
Compiling and running
Similar to the example programs, run
cmake -DDEAL_II_DIR=/path/to/deal.II .
in this directory to configure the problem.
You can switch between debug and release mode by calling either
or
The problem may then be run in serial mode with
and in parallel (in this case, on 6 processors) with
mpirun -np 6 ./nonlinear-poro-viscoelasticity
Alternatively, to keep it a bit more tidy, create a folder, e.g. run and copy the input file in there. Then type:
mpirun -np 6 -wdir run ../nonlinear-poro-viscoelasticity
All the input files used to produce the results shown in the paper are provided in in the input-files folder. Simply replace the parameters.prm file in the main directory. For the verification examples, run the python script 'run-multi-calc.py' instead:
The 'run-multi-calc.py' and 'runPoro.sh' files provided must both be in the main directory. This will automatically generate the required input files and run them in sequence.
Reference for this work
If you use this program as a basis for your own work, please consider citing the paper referenced in the introduction. The initial version of this work was contributed to the deal.II project by E. Comellas and J-P. Pelteret.
Recommended literature
-
W. Ehlers and G. Eipper (1999), Finite Elastic Deformations in Liquid-Saturated and Empty Porous Solids, Transport in Porous Media 34(1/3):179-191. DOI: 10.1023/A:1006565509095.
-
S. Reese and S. Govindjee (2001), A theory of finite viscoelasticity and numerical aspects, International Journal of Solids and Structures 35(26-27):3455-3482. DOI: 10.1016/S0020-7683(97)00217-5.
-
G. Franceschini, D. Bigoni, P. Regitnig and G. A. Holzapfel (2006), Brain tissue deforms similarly to filled elastomers and follows consolidation theory, Journal of the Mechanics and Physics of Solids 54(12):2592-2620. DOI: 10.1016/j.jmps.2006.05.004.
-
S. Budday, G. Sommer, J. Haybaeck, P. Steinmann, G. A. Holzapfel and E. Kuhl (2017), Rheological characterization of human brain tissue, Acta Biomaterialia 60:315-329. DOI: 10.1016/j.actbio.2017.06.024.
-
G.A. Holzapfel (2001), Nonlinear Solid Mechanics. A Continuum Approach for Engineering, John Wiley & Sons. ISBN: 978-0-471-82319-3.
Results
The results shown here are a selection of those presented and discussed in the paper referenced in the introduction.
Consolidation experiments
We reproduce the uniaxial consolidation experiments of brain tissue from the seminal paper by Franceschini et al. (2006) using a reduction of our formulation to nonlinear poroelasticity without the viscous component. The following geometry (320 cells and 9544 degrees of freedom), boundary conditions, and loading are used:
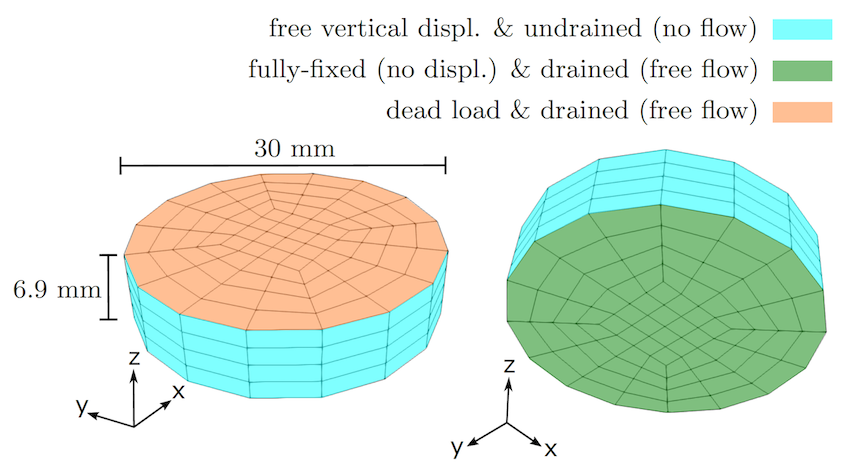
The material properties are:
-
First Lamé parameter: 334kPa
-
Ogden parameters: μ1=1.044kPa, μ2=1.183kPa, α1=4.309, and α2=7.736
-
Initial solid volume fraction: 0.80
-
Isotropic initial intrinsic permeability: 8.0e-11mm2
-
Deformation-dependency control parameter for the permeability: 40
-
Fluid viscosity: 0.89mPa·s
We consider the effect of gravity in our formulation, with the simplifying assumption that the fluid and solid density are both 0.997mg/mm3. Using these parameters and simulation conditions, we obtain a reasonable fit to the experimental curve:
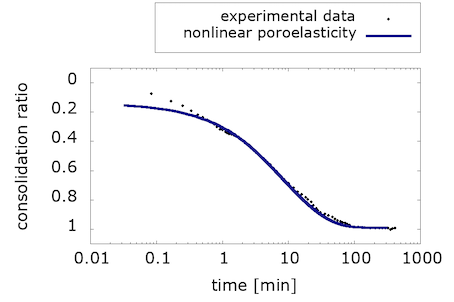
What is interesting about these results is that they show that nonlinear poroelasticity alone, i.e. with a purely hyperelastic solid component, can capture the consolidation behavior of brain tissue in response to an oedometric test. However, as we will see in the next section, the viscous component is necessary to also capture stress conditioning due to cyclic loading.
Cyclic experiments under multiple loading modes
Budday et al. (2017) performed an exhaustive set of cyclic loading experiments on brain tissue. They showed in this and subsequent publications that a finite viscoelastic Ogden model is able to predict essential features like nonlinearity, preconditioning, hysteresis, and tension-compression asymmetry. However, such a monophasic viscoelastic formulation can only implicitly capture the effects due to the extracellular fluid flow within the tissue. This was the motivation behind the formulation presented here: to provide a comprehensive model for brain tissue response that will enable us to explore the porous and viscous effects in its mechanical response to a wide range of loading scenarios.
As an example, we show here the results for cyclic compressive loading. The following geometry (512 cells and 15468 degrees of freedom), boundary conditions, and loading are used:
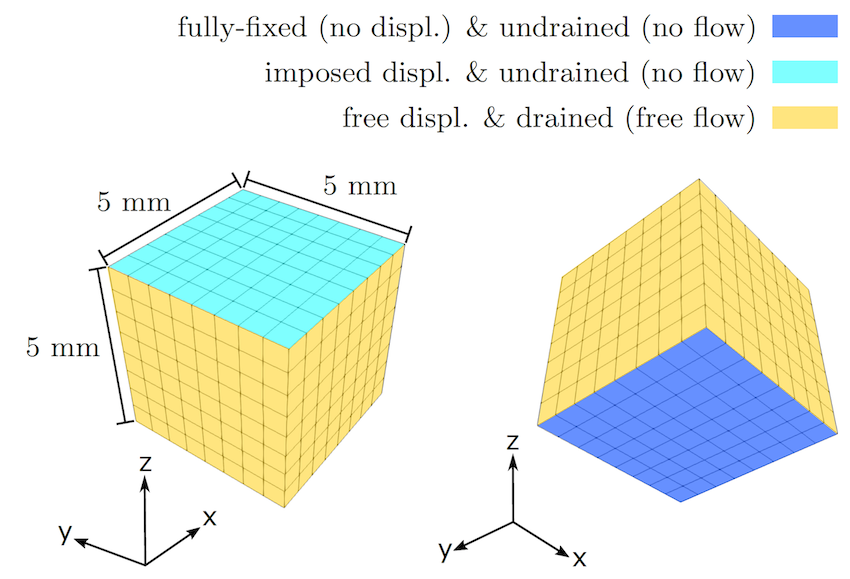
The material properties are:
-
First Lamé parameter: 24.5kPa
-
Ogden hyperelastic parameters: μ∞,1=-83.9Pa, and α∞,1=-11.92
-
Ogden viscous parameters: μ1=-2100Pa, and α1=-2.2
-
Deformation-independent viscosity of the solid: 14kPa·s
-
Initial solid volume fraction: 0.75
-
Isotropic initial intrinsic permeability: 1.0e-8mm2
-
Deformation-dependency control parameter for the permeability: 50
-
Fluid viscosity: 0.89mPa·s
To simplify the problem, we neglect the effect of gravity for this example. Here is an animation of our results, visualised with Paraview:

To compare with the experimental results, our code computes the nominal stress on the loading surface and provides the values for each timestep in the "data-for-gnuplot.sol" output file. We can then plot compressive nominal stress versus the averaged vertical stretch:
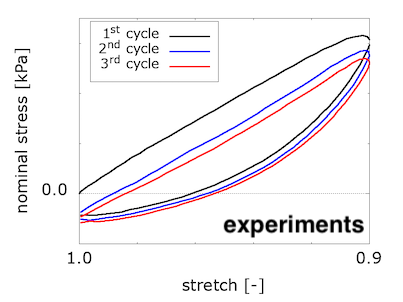
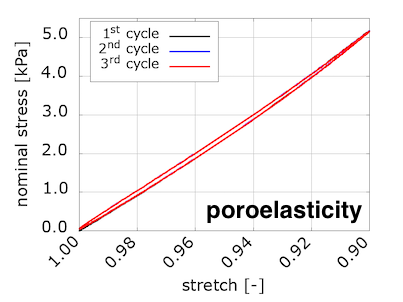
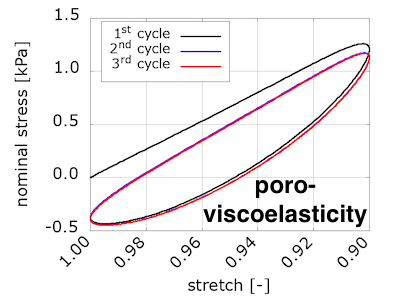
We see that viscosity is required in the solid component to reproduce the preconditioning and hysteretic response seen in the experiments. We were somewhat surprised by these results, because we were expecting to see preconditioning and hysteresis with poroelasticity alone. We discussed this in a conference talk:
-
E. Comellas, J.-P. Pelteret, S. Budday and P. Steinmann (2018). Unsolved issues in the numerical modelling of experimentally-observed porous effects in brain tissue behaviour, 6th European Conference on Computational Mechanics and 7th European Conference on Computational Fluid Dynamics (ECCM-ECFD 2018), Glasgow (UK), 11th–15th June 2018. DOI: 10.13140/RG.2.2.18553.29283.
So, we set out to explore why this is not the case. For that, we studied the porous and viscous dissipation, which are computed in the code and also provided in the "data-for-gnuplot.sol" output file. We determined that there is dissipation occurring in the poroelastic case, but we were barely seeing it in the stress-stretch plot. Even if we played around with the material parameters to increase the amount of dissipation, the slight hysteresis in our plot barely changed. Where is all that energy going?
We found the answer by looking closely into the thermodynamic basis of our constitutive equations. The viscous dissipation is a function of the viscous part of the stress tensor and the viscosity of the solid component. However, the porous dissipation depends on the porous material parameters and the seepage velocity, i.e. the relative velocity of the fluid with respect to the deforming solid component, which is proportional to the gradient of the pressure. Now, the plots we were studying are in the displacement-related space. We were looking for the porous dissipation in the wrong place! Our unknowns, the displacements and pressure, are slightly coupled. This is why we see a bit of porous dissipation in the stress-stretch plot for the poroelastic example.
Indeed, when we computed an equivalent to the nominal stress but in the pressure-related space, we found the "missing" dissipation. The "accumulated outgoing fluid volume" in the pressure-space is the equivalent to the nominal stress in the displacement-space. We plot it versus the fluid reaction pressure on the loading surface, which can be loosely seen as an equivalent to the averaged vertical stretch in the previous plots.
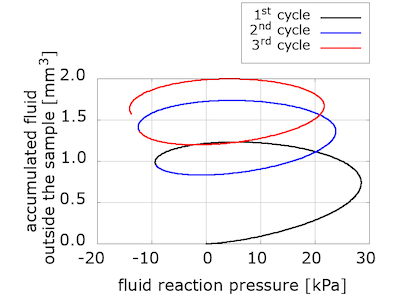
Our takeaways
In developing this formulation and running the examples provided in this code gallery, we have realised the importance of the loading and boundary conditions selected in the problems. Inhomogeneous displacements and pressure distributions require a fine enough discretization as well as a careful set-up of representative loading and boundary conditions. Without inhomogeneous stretch and pressure maps, the predicted viscous and porous behaviours of the material will not be realistic.
We have started to explore the relation between viscous and porous effects in response to different types of loading. Interestingly, the fluid accumulated outside the sample increases for each loading cycle in the poroelastic model, while it decreases for the poro-viscoelastic one.
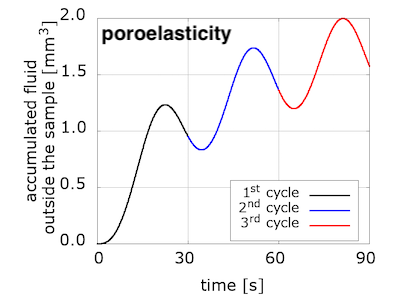
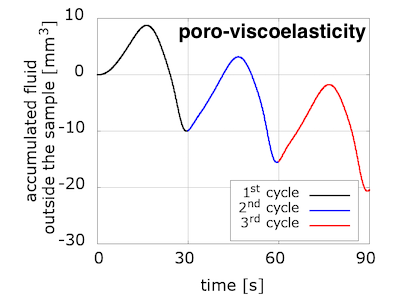
The evolution of pressure in the centre of the loading surface, where we observe a pressure concentration due to loading, has a noticeably different pattern for the two models. Note also the delay between the peak pressure values and peak displacement ones.
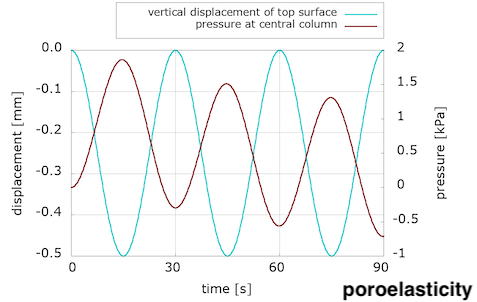
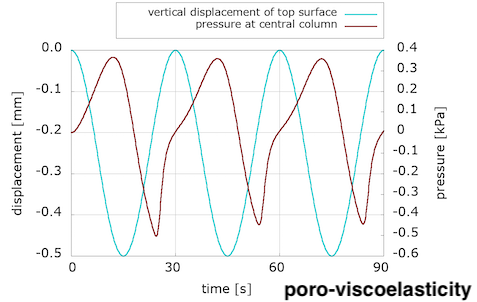
We are not sure yet how much of these differences is due to the incorporation of the viscous component, and how much can be explained by the choice of material parameters. (Note that the solid part of the poroelastic example has been adjusted with the Franceschini experiment, but the poro-viscoelastic one used the Budday experiments.) We will keep exploring with more numerical examples, but additional experiments are needed to adjust our material parameters in order to better distinguish the porous and viscous parts of brain tissue response to loading.
We hope the community finds this code useful, whether it is for brain mechanics or other applications, and look forward to learning and discussing about new insights gained about the interrelations of the viscous, porous, and elastic parts of the nonlinear poro-viscoelastic formulation.
Annotated version of input-files/5-Ehlers-verification/run-multi-calc.py
import fileinput
import sys
import subprocess
import os
import shutil
import select
import time
parallelProcesses = 1
processes = {}
jobnames = {}
x = open("parameters.prm", 'w')
x.write("subsection Finite element system\n")
x.write(" set Polynomial degree displ = 2\n")
x.write(" set Polynomial degree pore = 1\n")
x.write(" set Quadrature order = 3\n")
x.write("end\n\n")
x.write("subsection Geometry\n")
x.write(" set Geometry type = Ehlers_tube_step_load\n")
x.write(" set Global refinement = 1\n")
x.write(" set Grid scale = 1\n")
x.write(" set Load type = pressure\n")
x.write(" set Load value = -7.5e6\n")
x.write(" set Drained pressure = 0\n")
x.write("end\n\n")
x.write("subsection Material properties \n")
x.write(" set material = Neo-Hooke\n")
x.write(" set lambda = 8.375e6\n")
x.write(" set shear modulus = 5.583e6\n")
x.write(" set seepage definition = Ehlers\n")
x.write(" set initial solid volume fraction = 0.67\n")
x.write(" set kappa = 0\n")
x.write(" set initial Darcy coefficient = 1.0e-4\n")
x.write(" set fluid weight = 1.0e4\n")
x.write(" set gravity term = false\n")
x.write("end\n\n")
x.write("subsection Nonlinear solver\n")
x.write(" set Max iterations Newton-Raphson = 20\n")
x.write(" set Tolerance force = 1.0e-6\n")
x.write(" set Tolerance displacement = 1.0e-6\n")
x.write(" set Tolerance pore pressure = 1.0e-6\n")
x.write("end\n\n")
x.write("subsection Time\n")
x.write(" set End time = 10.0\n")
x.write(" set Time step size = 0.002\n")
x.write("end\n\n")
x.write("subsection Output parameters\n")
x.write(" set Time step number output = 5\n")
x.write("end\n")
x.close()
kappas = ["0","1","2","5"]
for kappa in kappas:
jobname = "Ehlers_step_load_kappa_"+kappa
x = fileinput.input("parameters.prm", inplace=1)
for line in x:
if "set kappa" in line:
line = " set kappa = "+kappa+"\n"
print (line),
x.close()
process = subprocess.Popen("./runPoro.sh " + jobname, shell=True)
executable = "RESULTS/calcDir_" + jobname
results = "RESULTS/resultDir_" + jobname
time.sleep(1)
while not os.path.exists(executable) and not os.path.exists(results):
time.sleep(1)
print ("waiting for executable to be copied")
processes[process.pid] = process
jobnames[process.pid] = jobname
while len(processes) >= parallelProcesses:
pid, status = os.wait()
if pid in processes:
if status == 0:
print ("Job %30s successful" % jobnames[pid])
del processes[pid]
del jobnames[pid]
while processes:
pid, status = os.wait()
if pid in processes:
if status == 0:
print ("Job %30s successful" % jobnames[pid])
del processes[pid]
del jobnames[pid]
jobname = "Ehlers_increase_load_kappa_0"
x = fileinput.input("parameters.prm", inplace=1)
for line in x:
if "set Geometry type" in line:
line = " set Geometry type = Ehlers_tube_increase_load\n"
if "set kappa" in line:
line = " set kappa = 0\n"
print (line),
x.close()
process = subprocess.Popen("./runPoro.sh " + jobname, shell=True)
executable = "RESULTS/calcDir_" + jobname
results = "RESULTS/resultDir_" + jobname
time.sleep(1)
while not os.path.exists(executable) and not os.path.exists(results):
time.sleep(1)
print ("waiting for executable to be copied")
processes[process.pid] = process
jobnames[process.pid] = jobname
while len(processes) >= parallelProcesses:
pid, status = os.wait()
if pid in processes:
if status == 0:
print ("Job %30s successful" % jobnames[pid])
del processes[pid]
del jobnames[pid]
jobname = "Ehlers_cube"
x = fileinput.input("parameters.prm", inplace=1)
for line in x:
if "set Geometry type" in line:
line = " set Geometry type = Ehlers_cube_consolidation\n"
if "set Global refinement" in line:
line = " set Global refinement = 2\n"
if "set Time step size" in line:
line = " set Time step size = 0.01\n"
if "set Time step number output" in line:
line = " set Time step number output = 1\n"
print (line),
x.close()
process = subprocess.Popen("./runPoro.sh " + jobname, shell=True)
executable = "RESULTS/calcDir_" + jobname
results = "RESULTS/resultDir_" + jobname
time.sleep(1)
while not os.path.exists(executable) and not os.path.exists(results):
time.sleep(1)
print ("waiting for executable to be copied")
processes[process.pid] = process
jobnames[process.pid] = jobname
while len(processes) >= parallelProcesses:
pid, status = os.wait()
if pid in processes:
if status == 0:
print ("Job %30s successful" % jobnames[pid])
del processes[pid]
del jobnames[pid]
print (" _ _ _ _ _ _ _ _ _ ")
print (" /_\ | | | (_)___| |__ ___ __ ___ _ __ _ __| |___| |_ ___ __| | |")
print (" / _ \| | | | / _ \ '_ (_-< / _/ _ \ ' \| '_ \ / -_) _/ -_) _ |_|")
print ("/_/ \_\_|_| _/ \___/_.__/__/ \__\___/_|_|_| .__/_\___|\__\___\__,_(_)")
print (" |__/ |_| ")
Annotated version of input-files/5-Ehlers-verification/runPoro.sh
#!/bin/bash
# first parameter is the name for directory where the calculation is made
echo " _ _ _ _ _ "
echo " _ _ _ _ _ _ _ _ (_)_ _ __ _ __| |___ __ _| | (_|_) "
echo "| '_| || | ' \| ' \| | ' \/ _ | / _ / -_) _ | |_| | | _ _ _ "
echo "|_| \_,_|_||_|_||_|_|_||_\__, | \__,_\___\__,_|_(_)_|_| (_) (_) (_) "
echo " |___/ "
echo "DEAL_II_DIR: " @f$DEAL_II_DIR
# Define and print tmpdir, where calculations are made and resultsdir where results will be stored
maindir=`pwd`
tmpdir=@f$maindir/RESULTS/calcDir_@f$1
resultdir=@f$maindir/RESULTS/resultDir_@f$1
echo "Main directory :" @f$maindir
echo "Directory for calculations:" @f$tmpdir
echo "Directory for results :" @f$resultdir
# change to temporary job directory
mkdir -p @f$tmpdir
cd @f$tmpdir
# copy stuff from location where job was submitted to temporary directory
cp -r @f$maindir/parameters.prm . && echo "Copying input file succeeded" || echo "Copying input file failed"
cp -r @f$maindir/nonlinear-poro-viscoelasticity . && echo "Copying executable succeeded" || echo "Copying executable failed"
# run code (change num of parallel processes if desired)
touch out.log
touch err.log
COUNTER=0
while [ @f$COUNTER -lt 10 ]; do
COUNTER=@f$((COUNTER+1))
echo "Start run " @f$COUNTER
mpirun -np 6 ./nonlinear-poro-viscoelasticity >>out.log 2>err.log
if [ @f$? -eq 0 ]; then
echo FEM Code OK
break
else
echo @f$?
echo FEM Code FAILED
fi
done
# create folder for output and copy parameter file and results into it
mkdir -p @f$resultdir
mkdir -p @f$resultdir/Paraview-files
cp parameters.prm @f$resultdir/
cp *.sol @f$resultdir/
cp solution.* @f$resultdir/Paraview-files/
# get rid of the temporary job dir
rm -rf @f$tmpdir
Annotated version of nonlinear-poro-viscoelasticity.cc
We start by including all the necessary deal.II header files and some C++ related ones. They have been discussed in detail in previous tutorial programs, so you need only refer to past tutorials for details.
#include <deal.II/base/function.h>
#include <deal.II/base/parameter_handler.h>
#include <deal.II/base/point.h>
#include <deal.II/base/quadrature_lib.h>
#include <deal.II/base/symmetric_tensor.h>
#include <deal.II/base/tensor.h>
#include <deal.II/base/timer.h>
#include <deal.II/base/work_stream.h>
#include <deal.II/base/mpi.h>
#include <deal.II/base/quadrature_point_data.h>
#include <deal.II/differentiation/ad.h>
#include <deal.II/distributed/shared_tria.h>
#include <deal.II/dofs/dof_renumbering.h>
#include <deal.II/dofs/dof_tools.h>
#include <deal.II/dofs/dof_accessor.h>
#include <deal.II/grid/filtered_iterator.h>
#include <deal.II/grid/grid_generator.h>
#include <deal.II/grid/grid_tools.h>
#include <deal.II/grid/grid_in.h>
#include <deal.II/grid/grid_out.h>
#include <deal.II/grid/manifold_lib.h>
#include <deal.II/grid/tria_accessor.h>
#include <deal.II/grid/tria_iterator.h>
#include <deal.II/fe/fe_dgp_monomial.h>
#include <deal.II/fe/fe_q.h>
#include <deal.II/fe/fe_system.h>
#include <deal.II/fe/fe_tools.h>
#include <deal.II/fe/fe_values.h>
#include <deal.II/lac/block_sparsity_pattern.h>
#include <deal.II/lac/affine_constraints.h>
#include <deal.II/lac/dynamic_sparsity_pattern.h>
#include <deal.II/lac/full_matrix.h>
#include <deal.II/lac/packaged_operation.h>
#include <deal.II/lac/trilinos_block_sparse_matrix.h>
#include <deal.II/lac/trilinos_linear_operator.h>
#include <deal.II/lac/trilinos_parallel_block_vector.h>
#include <deal.II/lac/trilinos_precondition.h>
#include <deal.II/lac/trilinos_sparse_matrix.h>
#include <deal.II/lac/trilinos_sparsity_pattern.h>
#include <deal.II/lac/trilinos_solver.h>
#include <deal.II/lac/trilinos_vector.h>
#include <deal.II/lac/block_vector.h>
#include <deal.II/lac/vector.h>
#include <deal.II/numerics/data_postprocessor.h>
#include <deal.II/numerics/data_out.h>
#include <deal.II/numerics/data_out_faces.h>
#include <deal.II/numerics/fe_field_function.h>
#include <deal.II/numerics/vector_tools.h>
#include <deal.II/physics/transformations.h>
#include <deal.II/physics/elasticity/kinematics.h>
#include <deal.II/physics/elasticity/standard_tensors.h>
#include <iostream>
#include <fstream>
#include <numeric>
#include <iomanip>
LinearOperator< Range, Domain, Payload > linear_operator(const OperatorExemplar &, const Matrix &)
We create a namespace for everything that relates to the nonlinear poro-viscoelastic formulation, and import all the deal.II function and class names into it:
namespace NonLinearPoroViscoElasticity
{
Run-time parameters
Set up a ParameterHandler object to read in the parameter choices at run-time introduced by the user through the file "parameters.prm"
Finite Element system
Here we specify the polynomial order used to approximate the solution, both for the displacements and pressure unknowns. The quadrature order should be adjusted accordingly.
{
unsigned int poly_degree_displ;
unsigned int poly_degree_pore;
unsigned int quad_order;
static void
void
};
{
{
"Displacement system polynomial order");
"Pore pressure system polynomial order");
"Gauss quadrature order");
}
}
{
{
poly_degree_displ = prm.
get_integer(
"Polynomial degree displ");
poly_degree_pore = prm.
get_integer(
"Polynomial degree pore");
}
}
void enter_subsection(const std::string &subsection, const bool create_path_if_needed=true)
long int get_integer(const std::string &entry_string) const
void declare_entry(const std::string &entry, const std::string &default_value, const Patterns::PatternBase &pattern=Patterns::Anything(), const std::string &documentation="", const bool has_to_be_set=false)
Geometry
These parameters are related to the geometry definition and mesh generation. We select the type of problem to solve and introduce the desired load values.
struct Geometry
{
std::string geom_type;
unsigned int global_refinement;
double scale;
std::string load_type;
double load;
unsigned int num_cycle_sets;
double fluid_flow;
double drained_pressure;
static void
void
};
{
{
"|Ehlers_tube_increase_load"
"|Ehlers_cube_consolidation"
"|Franceschini_consolidation"
"|Budday_cube_tension_compression"
"|Budday_cube_tension_compression_fully_fixed"
"|Budday_cube_shear_fully_fixed"),
"Type of geometry used. "
"For Ehlers verification examples see Ehlers and Eipper (1999). "
"For Franceschini brain consolidation see Franceschini et al. (2006)"
"For Budday brain examples see Budday et al. (2017)");
"Global refinement level");
"Global grid scaling factor");
"Type of loading");
"Loading value");
"Number of times each set of 3 cycles is repeated, only for "
"Budday_cube_tension_compression and Budday_cube_tension_compression_fully_fixed. "
"Load value is doubled in second set, load rate is kept constant."
"Final time indicates end of second cycle set.");
"Prescribed fluid flow. Not implemented in any example yet.");
"Increase of pressure value at drained boundary w.r.t the atmospheric pressure.");
}
}
{
{
geom_type = prm.
get(
"Geometry type");
global_refinement = prm.
get_integer(
"Global refinement");
load_type = prm.
get(
"Load type");
num_cycle_sets = prm.
get_integer(
"Number of cycle sets");
drained_pressure = prm.
get_double(
"Drained pressure");
}
}
std::string get(const std::string &entry_string) const
double get_double(const std::string &entry_name) const
Materials
Here we select the type of material for the solid component and define the corresponding material parameters. Then we define he fluid data, including the type of seepage velocity definition to use.
struct Materials
{
std::string mat_type;
double lambda;
double mu;
double mu1_infty;
double mu2_infty;
double mu3_infty;
double alpha1_infty;
double alpha2_infty;
double alpha3_infty;
double mu1_mode_1;
double mu2_mode_1;
double mu3_mode_1;
double alpha1_mode_1;
double alpha2_mode_1;
double alpha3_mode_1;
double viscosity_mode_1;
std::string fluid_type;
double solid_vol_frac;
double kappa_darcy;
double init_intrinsic_perm;
double viscosity_FR;
double init_darcy_coef;
double weight_FR;
bool gravity_term;
int gravity_direction;
double gravity_value;
double density_FR;
double density_SR;
static void
void
};
{
{
"Type of material used in the problem");
"First Lamé parameter for extension function related to compactation point in solid material [Pa].");
"shear modulus for Neo-Hooke materials [Pa].");
"The type of eigen solver to be used for Ogden and visco-Ogden models.");
"Shear material parameter 'mu1' for Ogden material [Pa].");
"Shear material parameter 'mu2' for Ogden material [Pa].");
"Shear material parameter 'mu1' for Ogden material [Pa].");
"Stiffness material parameter 'alpha1' for Ogden material [-].");
"Stiffness material parameter 'alpha2' for Ogden material [-].");
"Stiffness material parameter 'alpha3' for Ogden material [-].");
"Shear material parameter 'mu1' for first viscous mode in Ogden material [Pa].");
"Shear material parameter 'mu2' for first viscous mode in Ogden material [Pa].");
"Shear material parameter 'mu1' for first viscous mode in Ogden material [Pa].");
"Stiffness material parameter 'alpha1' for first viscous mode in Ogden material [-].");
"Stiffness material parameter 'alpha2' for first viscous mode in Ogden material [-].");
"Stiffness material parameter 'alpha3' for first viscous mode in Ogden material [-].");
"Deformation-independent viscosity parameter 'eta_1' for first viscous mode in Ogden material [-].");
"Type of formulation used to define the seepage velocity in the problem. "
"Choose between Markert formulation of deformation-dependent intrinsic permeability "
"and Ehlers formulation of deformation-dependent Darcy flow coefficient.");
"Initial porosity (solid volume fraction, 0 < n_0s < 1)");
"Deformation-dependency control parameter for specific permeability (kappa >= 0)");
"Initial intrinsic permeability parameter [m^2] (isotropic permeability). To be used with Markert formulation.");
"Effective shear viscosity parameter of the fluid [Pa·s, (N·s)/m^2]. To be used with Markert formulation.");
"Initial Darcy flow coefficient [m/s] (isotropic permeability). To be used with Ehlers formulation.");
"Effective weight of the fluid [N/m^3]. To be used with Ehlers formulation.");
"Gravity term considered (true) or neglected (false)");
"Real (or effective) density of the fluid");
"Real (or effective) density of the solid");
"Direction of gravity (unit vector 0 for x, 1 for y, 2 for z)");
"Value of gravity (be careful to have consistent units!)");
}
}
{
{
SymmetricTensorEigenvectorMethod
Solid
mat_type = prm.
get(
"material");
Fluid
fluid_type = prm.
get(
"seepage definition");
solid_vol_frac = prm.
get_double(
"initial solid volume fraction");
init_intrinsic_perm = prm.
get_double(
"initial intrinsic permeability");
init_darcy_coef = prm.
get_double(
"initial Darcy coefficient");
Gravity effects
gravity_term = prm.
get_bool(
"gravity term");
gravity_direction = prm.
get_integer(
"gravity direction");
if ( (fluid_type == "Markert") && ((init_intrinsic_perm == 0.0) || (viscosity_FR == 0.0)) )
AssertThrow(
false, ExcMessage(
"Markert seepage velocity formulation requires the definition of "
"'initial intrinsic permeability' and 'fluid viscosity' greater than 0.0."));
if ( (fluid_type == "Ehlers") && ((init_darcy_coef == 0.0) || (weight_FR == 0.0)) )
AssertThrow(
false, ExcMessage(
"Ehler seepage velocity formulation requires the definition of "
"'initial Darcy coefficient' and 'fluid weight' greater than 0.0."));
const std::string eigen_solver_type = prm.
get(
"eigen solver");
if (eigen_solver_type == "QL Implicit Shifts")
else if (eigen_solver_type == "Jacobi")
else
{
AssertThrow(
false, ExcMessage(
"Unknown eigen solver selected."));
}
}
}
bool get_bool(const std::string &entry_name) const
#define AssertThrow(cond, exc)
Nonlinear solver
We now define the tolerances and the maximum number of iterations for the Newton-Raphson scheme used to solve the nonlinear system of governing equations.
struct NonlinearSolver
{
unsigned int max_iterations_NR;
double tol_f;
double tol_u;
double tol_p_fluid;
static void
void
};
{
{
"Number of Newton-Raphson iterations allowed");
"Force residual tolerance");
"Displacement error tolerance");
"Pore pressure error tolerance");
}
}
{
{
max_iterations_NR = prm.
get_integer(
"Max iterations Newton-Raphson");
tol_p_fluid = prm.
get_double(
"Tolerance pore pressure");
}
}
Time
Here we set the timestep size \varDelta t and the simulation end-time.
struct Time
{
double end_time;
double delta_t;
static void
void
};
{
{
"End time");
"Time step size. The value must be larger than the displacement error tolerance defined.");
}
}
{
{
}
}
Output
We can choose the frequency of the data for the output files.
struct OutputParam
{
std::string outfiles_requested;
unsigned int timestep_output;
std::string outtype;
static void
void
};
{
{
"Paraview output files to generate.");
"Output data for time steps multiple of the given "
"integer value.");
"Output data associated with integration point values"
" averaged on elements or on nodes.");
}
}
{
{
outfiles_requested = prm.
get(
"Output files");
timestep_output = prm.
get_integer(
"Time step number output");
outtype = prm.
get(
"Averaged results");
}
}
All parameters
We finally consolidate all of the above structures into a single container that holds all the run-time selections.
public Geometry,
public Materials,
public NonlinearSolver,
public Time,
public OutputParam
{
AllParameters(const std::string &input_file);
static void
void
};
AllParameters::AllParameters(const std::string &input_file)
{
declare_parameters(prm);
prm.parse_input(input_file);
parse_parameters(prm);
}
{
FESystem::declare_parameters(prm);
Geometry::declare_parameters(prm);
Materials::declare_parameters(prm);
NonlinearSolver::declare_parameters(prm);
Time::declare_parameters(prm);
OutputParam::declare_parameters(prm);
}
{
FESystem::parse_parameters(prm);
Geometry::parse_parameters(prm);
Materials::parse_parameters(prm);
NonlinearSolver::parse_parameters(prm);
Time::parse_parameters(prm);
OutputParam::parse_parameters(prm);
}
}
Time class
A simple class to store time data. For simplicity we assume a constant time step size.
class Time
{
public:
Time (const double time_end,
const double delta_t)
:
timestep(0),
time_current(0.0),
time_end(time_end),
delta_t(delta_t)
{}
virtual ~Time()
{}
double get_current() const
{
return time_current;
}
double get_end() const
{
return time_end;
}
double get_delta_t() const
{
return delta_t;
}
unsigned int get_timestep() const
{
return timestep;
}
void increment_time ()
{
time_current += delta_t;
++timestep;
}
private:
unsigned int timestep;
double time_current;
double time_end;
const double delta_t;
};
Constitutive equation for the solid component of the biphasic material
Base class: generic hyperelastic material
The "extra" Kirchhoff stress in the solid component is the sum of isochoric and a volumetric part: \mathbf{\tau} = \mathbf{\tau}_E^{(\bullet)} + \mathbf{\tau}^{\textrm{vol}}. The deviatoric part changes depending on the type of material model selected: Neo-Hooken hyperelasticity, Ogden hyperelasticiy, or a single-mode finite viscoelasticity based on the Ogden hyperelastic model. In this base class we declare it as a virtual function, and it will be defined for each model type in the corresponding derived class. We define here the volumetric component, which depends on the extension function U(J_S) selected, and in this case is the same for all models. We use the function proposed by Ehlers & Eipper 1999 doi:10.1023/A:1006565509095. We also define some public functions to access and update the internal variables.
template <int dim, typename NumberType = Sacado::Fad::DFad<double> >
class Material_Hyperelastic
{
public:
Material_Hyperelastic(const Parameters::AllParameters ¶meters,
const Time &time)
:
n_OS (parameters.solid_vol_frac),
lambda (parameters.lambda),
time(time),
det_F (1.0),
det_F_converged (1.0),
eigen_solver (parameters.eigen_solver)
{}
~Material_Hyperelastic()
{}
get_tau_E(const
Tensor<2,dim, NumberType> &F) const
{
return ( get_tau_E_base(F) + get_tau_E_ext_func(F) );
}
{
Assert(det_F > 0, ExcInternalError());
return get_tau_E(F)*NumberType(1/det_F);
}
double
get_converged_det_F() const
{
return det_F_converged;
}
virtual void
update_end_timestep()
{
det_F_converged = det_F;
}
virtual void
{
}
virtual double
get_viscous_dissipation( ) const = 0;
const double n_OS;
const Time &time;
double det_F;
double det_F_converged;
protected:
{
Assert(det_F > 0, ExcInternalError());
return ( NumberType(lambda * (1.0-n_OS)*(1.0-n_OS)
* (det_F/(1.0-n_OS) - det_F/(det_F-n_OS))) * I );
}
};
#define Assert(cond, exc)
DEAL_II_HOST constexpr Number determinant(const SymmetricTensor< 2, dim, Number > &)
Derived class: Neo-Hookean hyperelastic material
template <int dim, typename NumberType = Sacado::Fad::DFad<double> >
class NeoHooke : public Material_Hyperelastic < dim, NumberType >
{
public:
NeoHooke(const Parameters::AllParameters ¶meters,
const Time &time)
:
Material_Hyperelastic< dim, NumberType > (parameters,time),
mu(parameters.mu)
{}
virtual ~NeoHooke()
{}
double
get_viscous_dissipation() const override
{
return 0.0;
}
protected:
const double mu;
{
const bool use_standard_model = true;
if (use_standard_model)
{
Standard Neo-Hooke
}
else
{
DEAL_II_HOST constexpr SymmetricTensor< 2, dim, Number > symmetrize(const Tensor< 2, dim, Number > &t)
Neo-Hooke in terms of principal stretches
const std::array< std::pair< NumberType, Tensor< 1, dim, NumberType > >, dim >
for (unsigned int d=0; d<dim; ++d)
return ( mu*(B_ev-I) );
}
}
};
DEAL_II_HOST constexpr SymmetricTensor< 4, dim, Number > outer_product(const SymmetricTensor< 2, dim, Number > &t1, const SymmetricTensor< 2, dim, Number > &t2)
std::array< std::pair< Number, Tensor< 1, dim, Number > >, std::integral_constant< int, dim >::value > eigenvectors(const SymmetricTensor< 2, dim, Number > &T, const SymmetricTensorEigenvectorMethod method=SymmetricTensorEigenvectorMethod::ql_implicit_shifts)
Derived class: Ogden hyperelastic material
template <int dim, typename NumberType = Sacado::Fad::DFad<double> >
class Ogden : public Material_Hyperelastic < dim, NumberType >
{
public:
Ogden(const Parameters::AllParameters ¶meters,
const Time &time)
:
Material_Hyperelastic< dim, NumberType > (parameters,time),
mu({parameters.mu1_infty,
parameters.mu2_infty,
parameters.mu3_infty}),
alpha({parameters.alpha1_infty,
parameters.alpha2_infty,
parameters.alpha3_infty})
{}
virtual ~Ogden()
{}
double
get_viscous_dissipation() const override
{
return 0.0;
}
protected:
std::vector<double> mu;
std::vector<double> alpha;
{
const std::array< std::pair< NumberType, Tensor< 1, dim, NumberType > >, dim >
for (unsigned int i = 0; i < 3; ++i)
{
for (unsigned int A = 0; A < dim; ++A)
{
tau += tau_aux1;
}
tau_aux2 *= mu[i];
tau -= tau_aux2;
}
return tau;
}
};
::VectorizedArray< Number, width > pow(const ::VectorizedArray< Number, width > &, const Number p)
Derived class: Single-mode Ogden viscoelastic material
We use the finite viscoelastic model described in Reese & Govindjee (1998) doi:10.1016/S0020-7683(97)00217-5 The algorithm for the implicit exponential time integration is given in Budday et al. (2017) doi: 10.1016/j.actbio.2017.06.024
template <int dim, typename NumberType = Sacado::Fad::DFad<double> >
class visco_Ogden : public Material_Hyperelastic < dim, NumberType >
{
public:
visco_Ogden(const Parameters::AllParameters ¶meters,
const Time &time)
:
Material_Hyperelastic< dim, NumberType > (parameters,time),
mu_infty({parameters.mu1_infty,
parameters.mu2_infty,
parameters.mu3_infty}),
alpha_infty({parameters.alpha1_infty,
parameters.alpha2_infty,
parameters.alpha3_infty}),
mu_mode_1({parameters.mu1_mode_1,
parameters.mu2_mode_1,
parameters.mu3_mode_1}),
alpha_mode_1({parameters.alpha1_mode_1,
parameters.alpha2_mode_1,
parameters.alpha3_mode_1}),
viscosity_mode_1(parameters.viscosity_mode_1),
{}
virtual ~visco_Ogden()
{}
void
{
Material_Hyperelastic < dim, NumberType >::update_internal_equilibrium(
F);
this->Cinv_v_1 = this->Cinv_v_1_converged;
const std::array< std::pair< NumberType, Tensor< 1, dim, NumberType > >, dim >
eigen_B_e_1_tr =
eigenvectors(B_e_1_tr, this->eigen_solver);
for (int a = 0; a < dim; ++a)
{
epsilon_e_1_tr[a] =
std::log(lambdas_e_1_tr[a]);
}
const double tolerance = 1
e-8;
double residual_check = tolerance*10.0;
std::vector<NumberType> lambdas_e_1_iso(dim);
int iteration = 0;
epsilon_e_1 = epsilon_e_1_tr;
while(residual_check > tolerance)
{
NumberType aux_J_e_1 = 1.0;
for (unsigned int a = 0; a < dim; ++a)
{
lambdas_e_1[a] =
std::exp(epsilon_e_1[a]);
aux_J_e_1 *= lambdas_e_1[a];
}
J_e_1 = aux_J_e_1;
for (unsigned int a = 0; a < dim; ++a)
lambdas_e_1_iso[a] = lambdas_e_1[a]*
std::pow(J_e_1,-1.0/dim);
for (unsigned int a = 0; a < dim; ++a)
{
residual[a] = get_beta_mode_1(lambdas_e_1_iso, a);
residual[a] *= this->time.get_delta_t()/(2.0*viscosity_mode_1);
residual[a] += epsilon_e_1[a];
residual[a] -= epsilon_e_1_tr[a];
for (
unsigned int b = 0;
b < dim; ++
b)
{
tangent[a][
b] = get_gamma_mode_1(lambdas_e_1_iso, a, b);
tangent[a][
b] *= this->time.get_delta_t()/(2.0*viscosity_mode_1);
tangent[a][
b] += I[a][
b];
}
}
epsilon_e_1 -=
invert(tangent)*residual;
residual_check = 0.0;
for (unsigned int a = 0; a < dim; ++a)
{
if (
std::abs(residual[a]) > residual_check)
}
iteration += 1;
if (iteration > 15 )
AssertThrow(
false, ExcMessage(
"No convergence in local Newton iteration for the "
"viscoelastic exponential time integration algorithm."));
}
NumberType aux_J_e_1 = 1.0;
for (unsigned int a = 0; a < dim; ++a)
{
lambdas_e_1[a] =
std::exp(epsilon_e_1[a]);
aux_J_e_1 *= lambdas_e_1[a];
}
J_e_1 = aux_J_e_1;
for (unsigned int a = 0; a < dim; ++a)
lambdas_e_1_iso[a] = lambdas_e_1[a]*
std::pow(J_e_1,-1.0/dim);
for (unsigned int a = 0; a < dim; ++a)
{
B_e_1_aux *= lambdas_e_1[a] * lambdas_e_1[a];
B_e_1 += B_e_1_aux;
}
this->tau_neq_1 = 0;
for (unsigned int a = 0; a < dim; ++a)
{
tau_neq_1_aux *= get_beta_mode_1(lambdas_e_1_iso, a);
this->tau_neq_1 += tau_neq_1_aux;
}
SymmetricTensor< 2, dim, Number > e(const Tensor< 2, dim, Number > &F)
SymmetricTensor< 2, dim, Number > b(const Tensor< 2, dim, Number > &F)
Tensor< 2, dim, Number > F(const Tensor< 2, dim, Number > &Grad_u)
::VectorizedArray< Number, width > log(const ::VectorizedArray< Number, width > &)
::VectorizedArray< Number, width > exp(const ::VectorizedArray< Number, width > &)
::VectorizedArray< Number, width > sqrt(const ::VectorizedArray< Number, width > &)
::VectorizedArray< Number, width > abs(const ::VectorizedArray< Number, width > &)
DEAL_II_HOST constexpr SymmetricTensor< 2, dim, Number > invert(const SymmetricTensor< 2, dim, Number > &)
Store history
for (unsigned int a = 0; a < dim; ++a)
for (unsigned int b = 0; b < dim; ++b)
}
void update_end_timestep() override
{
Material_Hyperelastic < dim, NumberType >::update_end_timestep();
this->Cinv_v_1_converged = this->Cinv_v_1;
}
double get_viscous_dissipation() const override
{
NumberType dissipation_term = get_tau_E_neq() * get_tau_E_neq();
dissipation_term /= (2*viscosity_mode_1);
return dissipation_term.val();
}
protected:
std::vector<double> mu_infty;
std::vector<double> alpha_infty;
std::vector<double> mu_mode_1;
std::vector<double> alpha_mode_1;
double viscosity_mode_1;
{
return ( get_tau_E_neq() + get_tau_E_eq(F) );
}
{
std::array< std::pair< NumberType, Tensor< 1, dim, NumberType > >, dim > eigen_B;
for (unsigned int i = 0; i < 3; ++i)
{
for (unsigned int A = 0; A < dim; ++A)
{
tau_aux1 *= mu_infty[i]*
std::pow(eigen_B[A].
first, (alpha_infty[i]/2.) );
tau += tau_aux1;
}
tau_aux2 *= mu_infty[i];
tau -= tau_aux2;
}
return tau;
}
get_tau_E_neq() const
{
return tau_neq_1;
}
NumberType
get_beta_mode_1(std::vector< NumberType > &lambda, const int &A) const
{
NumberType beta = 0.0;
for (unsigned int i = 0; i < 3; ++i)
{
NumberType aux = 0.0;
for (int p = 0; p < dim; ++p)
aux +=
std::pow(lambda[p],alpha_mode_1[i]);
aux *= -1.0/dim;
aux +=
std::pow(lambda[A], alpha_mode_1[i]);
aux *= mu_mode_1[i];
beta += aux;
}
return beta;
}
NumberType
get_gamma_mode_1(std::vector< NumberType > &lambda,
const int &A,
const int &B ) const
{
if (A==B)
{
for (unsigned int i = 0; i < 3; ++i)
{
NumberType aux = 0.0;
for (int p = 0; p < dim; ++p)
aux +=
std::pow(lambda[p],alpha_mode_1[i]);
aux *= 1.0/(dim*dim);
aux += 1.0/dim *
std::pow(lambda[A], alpha_mode_1[i]);
aux *= mu_mode_1[i]*alpha_mode_1[i];
}
}
else
{
for (unsigned int i = 0; i < 3; ++i)
{
NumberType aux = 0.0;
for (int p = 0; p < dim; ++p)
aux +=
std::pow(lambda[p],alpha_mode_1[i]);
aux *= 1.0/(dim*dim);
aux -= 1.0/dim *
std::pow(lambda[A], alpha_mode_1[i]);
aux -= 1.0/dim *
std::pow(lambda[B], alpha_mode_1[i]);
aux *= mu_mode_1[i]*alpha_mode_1[i];
}
}
}
};
long double gamma(const unsigned int n)
Constitutive equation for the fluid component of the biphasic material
We consider two slightly different definitions to define the seepage velocity with a Darcy-like law. Ehlers & Eipper 1999, doi:10.1023/A:1006565509095 Markert 2007, doi:10.1007/s11242-007-9107-6 The selection of one or another is made by the user via the parameters file.
template <int dim, typename NumberType = Sacado::Fad::DFad<double> >
class Material_Darcy_Fluid
{
public:
Material_Darcy_Fluid(const Parameters::AllParameters ¶meters)
:
fluid_type(parameters.fluid_type),
n_OS(parameters.solid_vol_frac),
initial_intrinsic_permeability(parameters.init_intrinsic_perm),
viscosity_FR(parameters.viscosity_FR),
initial_darcy_coefficient(parameters.init_darcy_coef),
weight_FR(parameters.weight_FR),
kappa_darcy(parameters.kappa_darcy),
gravity_term(parameters.gravity_term),
density_FR(parameters.density_FR),
gravity_direction(parameters.gravity_direction),
gravity_value(parameters.gravity_value)
{
Assert(kappa_darcy >= 0, ExcInternalError());
}
~Material_Darcy_Fluid()
{}
{
Assert(det_F > 0.0, ExcInternalError());
if (fluid_type == "Markert")
permeability_term = get_instrinsic_permeability_current(F) / viscosity_FR;
else if (fluid_type == "Ehlers")
permeability_term = get_darcy_flow_current(F) / weight_FR;
else
"Material_Darcy_Fluid --> Only Markert "
"and Ehlers formulations have been implemented."));
return ( -1.0 * permeability_term * det_F
* (grad_p_fluid - get_body_force_FR_current()) );
}
{
NumberType dissipation_term;
Assert(det_F > 0.0, ExcInternalError());
if (fluid_type == "Markert")
{
permeability_term = get_instrinsic_permeability_current(F) / viscosity_FR;
seepage_velocity = get_seepage_velocity_current(F,grad_p_fluid);
}
else if (fluid_type == "Ehlers")
{
permeability_term = get_darcy_flow_current(F) / weight_FR;
seepage_velocity = get_seepage_velocity_current(F,grad_p_fluid);
}
else
"Material_Darcy_Fluid --> Only Markert and Ehlers "
"formulations have been implemented."));
dissipation_term = (
invert(permeability_term) * seepage_velocity ) * seepage_velocity;
dissipation_term *= 1.0/(det_F*det_F);
}
protected:
const std::string fluid_type;
const double n_OS;
const double initial_intrinsic_permeability;
const double viscosity_FR;
const double initial_darcy_coefficient;
const double weight_FR;
const double kappa_darcy;
const bool gravity_term;
const double density_FR;
const int gravity_direction;
const double gravity_value;
{
Assert(det_F > 0.0, ExcInternalError());
const NumberType fraction = (det_F - n_OS)/(1 - n_OS);
return ( NumberType (
std::pow(fraction, kappa_darcy))
* initial_instrinsic_permeability_tensor );
}
{
Assert(det_F > 0.0, ExcInternalError());
const NumberType fraction = (1.0 - (n_OS / det_F) )/(1.0 - n_OS);
return ( NumberType (
std::pow(fraction, kappa_darcy))
* initial_darcy_flow_tensor);
}
get_body_force_FR_current() const
{
if (gravity_term == true)
{
gravity_vector[gravity_direction] = gravity_value;
body_force_FR_current = density_FR * gravity_vector;
}
return body_force_FR_current;
}
};
As seen in step-18, the PointHistory class offers a method for storing data at the quadrature points. Here each quadrature point holds a pointer to a material description. Thus, different material models can be used in different regions of the domain. Among other data, we choose to store the "extra" Kirchhoff stress \boldsymbol{\tau}_E and the dissipation values \mathcal{D}_p and \mathcal{D}_v.
template <int dim, typename NumberType = Sacado::Fad::DFad<double> >
class PointHistory
{
public:
PointHistory()
{}
virtual ~PointHistory()
{}
void setup_lqp (const Parameters::AllParameters ¶meters,
const Time &time)
{
if (parameters.mat_type == "Neo-Hooke")
solid_material.reset(new NeoHooke<dim,NumberType>(parameters,time));
else if (parameters.mat_type == "Ogden")
solid_material.reset(new Ogden<dim,NumberType>(parameters,time));
else if (parameters.mat_type == "visco-Ogden")
solid_material.reset(new visco_Ogden<dim,NumberType>(parameters,time));
else
Assert (
false, ExcMessage(
"Material type not implemented"));
fluid_material.reset(new Material_Darcy_Fluid<dim,NumberType>(parameters));
}
{
return solid_material->get_tau_E(F);
}
{
return solid_material->get_Cauchy_E(F);
}
double
get_converged_det_F() const
{
return solid_material->get_converged_det_F();
}
void
update_end_timestep()
{
solid_material->update_end_timestep();
}
void
{
solid_material->update_internal_equilibrium(F);
}
double
get_viscous_dissipation() const
{
return solid_material->get_viscous_dissipation();
}
{
return fluid_material->get_seepage_velocity_current(F, grad_p_fluid);
}
double
{
return fluid_material->get_porous_dissipation(F, grad_p_fluid);
}
const Parameters::AllParameters ¶meters) const
{
if (parameters.gravity_term == true)
{
Assert(det_F_AD > 0.0, ExcInternalError());
const NumberType overall_density_ref
= parameters.density_SR * parameters.solid_vol_frac
+ parameters.density_FR
* (det_F_AD - parameters.solid_vol_frac);
gravity_vector[parameters.gravity_direction] = parameters.gravity_value;
body_force = overall_density_ref * gravity_vector;
}
return body_force;
}
private:
std::shared_ptr< Material_Hyperelastic<dim, NumberType> > solid_material;
std::shared_ptr< Material_Darcy_Fluid<dim, NumberType> > fluid_material;
};
Nonlinear poro-viscoelastic solid
The Solid class is the central class as it represents the problem at hand: the nonlinear poro-viscoelastic solid
template <int dim>
class Solid
{
public:
Solid(const Parameters::AllParameters ¶meters);
virtual ~Solid();
void run();
protected:
using ADNumberType = Sacado::Fad::DFad<double>;
std::ofstream outfile;
std::ofstream pointfile;
struct PerTaskData_ASM;
template<typename NumberType = double> struct ScratchData_ASM;
Generate mesh
virtual void make_grid() = 0;
Define points for post-processing
virtual void define_tracked_vertices(std::vector<
Point<dim> > &tracked_vertices) = 0;
Set up the finite element system to be solved:
Extract sub-blocks from the global matrix
void determine_component_extractors();
Several functions to assemble the system and right hand side matrices using multithreading.
void assemble_system
void assemble_system_one_cell
ScratchData_ASM<ADNumberType> &scratch,
PerTaskData_ASM &data) const;
void copy_local_to_global_system(const PerTaskData_ASM &data);
typename ActiveSelector::active_cell_iterator active_cell_iterator
Define boundary conditions
virtual void make_constraints(const int &it_nr);
virtual double get_prescribed_fluid_flow
get_reaction_boundary_id_for_output () const = 0;
virtual std::pair<types::boundary_id,types::boundary_id>
get_drained_boundary_id_for_output () const = 0;
virtual std::vector<double> get_dirichlet_load
const int &direction) const = 0;
Create and update the quadrature points.
Solve non-linear system using a Newton-Raphson scheme
Solve the linearized equations using a direct solver
Retrieve the solution
Store the converged values of the internal variables at the end of each timestep
void update_end_timestep();
Post-processing and writing data to files
void output_results_to_vtu(const unsigned int timestep,
const double current_time,
void output_results_to_plot(const unsigned int timestep,
const double current_time,
std::ofstream &pointfile) const;
Headers and footer for the output files
void print_console_file_header( std::ofstream &outfile) const;
void print_plot_file_header(std::vector<
Point<dim> > &tracked_vertices,
std::ofstream &pointfile) const;
void print_console_file_footer(std::ofstream &outfile) const;
void print_plot_file_footer( std::ofstream &pointfile) const;
For parallel communication
const unsigned int n_mpi_processes;
const unsigned int this_mpi_process;
A collection of the parameters used to describe the problem setup
const Parameters::AllParameters ¶meters;
Declare an instance of dealii Triangulation class (mesh)
const ::parallel::distributed::Triangulation< dim, spacedim > * triangulation
Keep track of the current time and the time spent evaluating certain functions
A storage object for quadrature point information.
Integers to store polynomial degree (needed for output)
const unsigned int degree_displ;
const unsigned int degree_pore;
Declare an instance of dealii FESystem class (finite element definition)
Declare an instance of dealii DoFHandler class (assign DoFs to mesh)
Integer to store DoFs per element (this value will be used often)
const unsigned int dofs_per_cell;
Declare an instance of dealii Extractor objects used to retrieve information from the solution vectors We will use "u_fe" and "p_fluid_fe"as subscript in operator [] expressions on FEValues and FEFaceValues objects to extract the components of the displacement vector and fluid pressure, respectively.
Description of how the block-system is arranged. There are 3 blocks: 0 - vector DOF displacements u 1 - scalar DOF fluid pressure p_fluid
static const unsigned int n_blocks = 2;
static const unsigned int n_components = dim+1;
static const unsigned int first_u_component = 0;
static const unsigned int p_fluid_component = dim;
enum
{
u_block = 0,
p_fluid_block = 1
};
Extractors
Block data
std::vector<unsigned int> block_component;
DoF index data
std::vector<IndexSet> all_locally_owned_dofs;
std::vector<IndexSet> locally_owned_partitioning;
std::vector<IndexSet> locally_relevant_partitioning;
std::vector<types::global_dof_index> dofs_per_block;
std::vector<types::global_dof_index> element_indices_u;
std::vector<types::global_dof_index> element_indices_p_fluid;
Declare an instance of dealii QGauss class (The Gauss-Legendre family of quadrature rules for numerical integration) Gauss Points in element, with n quadrature points (in each space direction <dim> )
Gauss Points on element faces (used for definition of BCs)
const QGauss<dim - 1> qf_face;
Integer to store num GPs per element (this value will be used often)
const unsigned int n_q_points;
Integer to store num GPs per face (this value will be used often)
const unsigned int n_q_points_f;
Declare an instance of dealii AffineConstraints class (linear constraints on DoFs due to hanging nodes or BCs)
Declare an instance of dealii classes necessary for FE system set-up and assembly Store elements of tangent matrix (indicated by SparsityPattern class) as sparse matrix (more efficient)
Right hand side vector of forces
Total displacement values + pressure (accumulated solution to FE system)
Non-block system for the direct solver. We will copy the block system into these to solve the linearized system of equations.
We define variables to store norms and update norms and normalisation factors.
struct Errors
{
Errors()
:
norm(1.0), u(1.0), p_fluid(1.0)
{}
void reset()
{
norm = 1.0;
u = 1.0;
p_fluid = 1.0;
}
void normalise(const Errors &rhs)
{
if (rhs.norm != 0.0)
if (rhs.u != 0.0)
u /= rhs.u;
if (rhs.p_fluid != 0.0)
p_fluid /= rhs.p_fluid;
}
};
double norm(const FEValuesBase< dim > &fe, const ArrayView< const std::vector< Tensor< 1, dim > > > &Du)
Declare several instances of the "Error" structure
Errors error_residual, error_residual_0, error_residual_norm, error_update,
error_update_0, error_update_norm;
Methods to calculate error measures
void get_error_residual(Errors &error_residual_OUT);
void get_error_update
Errors &error_update_OUT);
Print information to screen
void print_conv_header();
void print_conv_footer();
NOTE: In all functions, we pass by reference (&), so these functions work on the original copy (not a clone copy), modifying the input variables inside the functions will change them outside the function.
Implementation of the Solid class
Public interface
We initialise the Solid class using data extracted from the parameter file.
template <int dim>
Solid<dim>::Solid(const Parameters::AllParameters ¶meters)
:
mpi_communicator(MPI_COMM_WORLD),
n_mpi_processes (
Utilities::
MPI::n_mpi_processes(mpi_communicator)),
this_mpi_process (
Utilities::
MPI::this_mpi_process(mpi_communicator)),
pcout(
std::cout, this_mpi_process == 0),
parameters(parameters),
time(parameters.end_time, parameters.delta_t),
timerconsole( mpi_communicator,
pcout,
timerfile( mpi_communicator,
outfile,
degree_displ(parameters.poly_degree_displ),
degree_pore(parameters.poly_degree_pore),
fe(
FE_Q<dim>(parameters.poly_degree_displ), dim,
FE_Q<dim>(parameters.poly_degree_pore), 1 ),
dofs_per_cell (fe.dofs_per_cell),
u_fe(first_u_component),
p_fluid_fe(p_fluid_component),
x_displacement(first_u_component),
y_displacement(first_u_component+1),
z_displacement(first_u_component+2),
pressure(p_fluid_component),
dofs_per_block(n_blocks),
qf_cell(parameters.quad_order),
qf_face(parameters.quad_order),
n_q_points (qf_cell.size()),
n_q_points_f (qf_face.size())
{
Assert(dim==3, ExcMessage(
"This problem only works in 3 space dimensions."));
determine_component_extractors();
}
The class destructor simply clears the data held by the DOFHandler
template <int dim>
Solid<dim>::~Solid()
{
dof_handler_ref.clear();
}
Runs the 3D solid problem
template <int dim>
void Solid<dim>::run()
{
The current solution increment is defined as a block vector to reflect the structure of the PDE system, with multiple solution components
Open file
if (this_mpi_process == 0)
{
outfile.open("console-output.sol");
print_console_file_header(outfile);
}
Generate mesh
Assign DOFs and create the stiffness and right-hand-side force vector
system_setup(solution_delta);
Define points for post-processing
std::vector<Point<dim> > tracked_vertices (2);
define_tracked_vertices(tracked_vertices);
std::vector<Point<dim>> reaction_force;
if (this_mpi_process == 0)
{
pointfile.open("data-for-gnuplot.sol");
print_plot_file_header(tracked_vertices, pointfile);
}
Print results to output file
if (parameters.outfiles_requested == "true")
{
output_results_to_vtu(time.get_timestep(),
time.get_current(),
solution_n );
}
output_results_to_plot(time.get_timestep(),
time.get_current(),
solution_n,
tracked_vertices,
pointfile);
Increment time step (=load step) NOTE: In solving the quasi-static problem, the time becomes a loading parameter, i.e. we increase the loading linearly with time, making the two concepts interchangeable.
Print information on screen
pcout << "\nSolver:";
pcout << "\n CST = make constraints";
pcout << "\n ASM_SYS = assemble system";
pcout << "\n SLV = linear solver \n";
Print information on file
outfile << "\nSolver:";
outfile << "\n CST = make constraints";
outfile << "\n ASM_SYS = assemble system";
outfile << "\n SLV = linear solver \n";
while ( (time.get_end() - time.get_current()) > -1.0*parameters.tol_u )
{
Initialize the current solution increment to zero
Solve the non-linear system using a Newton-Rapshon scheme
solve_nonlinear_timestep(solution_delta);
Add the computed solution increment to total solution
solution_n += solution_delta;
Store the converged values of the internal variables
Output results
if (( (time.get_timestep()%parameters.timestep_output) == 0 )
&& (parameters.outfiles_requested == "true") )
{
output_results_to_vtu(time.get_timestep(),
time.get_current(),
solution_n );
}
output_results_to_plot(time.get_timestep(),
time.get_current(),
solution_n,
tracked_vertices,
pointfile);
Increment the time step (=load step)
Print the footers and close files
if (this_mpi_process == 0)
{
print_plot_file_footer(pointfile);
pointfile.close ();
print_console_file_footer(outfile);
NOTE: ideally, we should close the outfile here [ >> outfile.close (); ] But if we do, then the timer output will not be printed. That is why we leave it open.
Private interface
We define the structures needed for parallelization with Threading Building Blocks (TBB) Tangent matrix and right-hand side force vector assembly structures. PerTaskData_ASM stores local contributions
template <int dim>
struct Solid<dim>::PerTaskData_ASM
{
std::vector<types::global_dof_index> local_dof_indices;
PerTaskData_ASM(const unsigned int dofs_per_cell)
:
cell_matrix(dofs_per_cell, dofs_per_cell),
cell_rhs(dofs_per_cell),
local_dof_indices(dofs_per_cell)
{}
void reset()
{
cell_matrix = 0.0;
cell_rhs = 0.0;
}
};
ScratchData_ASM stores larger objects used during the assembly
template <int dim>
template <typename NumberType>
struct Solid<dim>::ScratchData_ASM
{
Integration helper
Quadrature point solution
std::vector<NumberType> local_dof_values;
std::vector<Tensor<2, dim, NumberType> > solution_grads_u_total;
std::vector<NumberType> solution_values_p_fluid_total;
std::vector<Tensor<1, dim, NumberType> > solution_grads_p_fluid_total;
std::vector<Tensor<1, dim, NumberType> > solution_grads_face_p_fluid_total;
shape function values
std::vector<std::vector<Tensor<1,dim>>> Nx;
std::vector<std::vector<double>> Nx_p_fluid;
shape function gradients
std::vector<std::vector<Tensor<2,dim, NumberType>>> grad_Nx;
std::vector<std::vector<SymmetricTensor<2,dim, NumberType>>> symm_grad_Nx;
std::vector<std::vector<Tensor<1,dim, NumberType>>> grad_Nx_p_fluid;
:
solution_total (solution_total),
fe_values_ref(fe_cell, qf_cell, uf_cell),
fe_face_values_ref(fe_cell, qf_face, uf_face),
local_dof_values(fe_cell.dofs_per_cell),
solution_grads_u_total(qf_cell.size()),
solution_values_p_fluid_total(qf_cell.size()),
solution_grads_p_fluid_total(qf_cell.size()),
solution_grads_face_p_fluid_total(qf_face.size()),
Nx(qf_cell.size(),
std::vector<
Tensor<1,dim>>(fe_cell.dofs_per_cell)),
Nx_p_fluid(qf_cell.size(),
std::vector<double>(fe_cell.dofs_per_cell)),
grad_Nx(qf_cell.size(),
std::vector<
Tensor<2, dim, NumberType>>(fe_cell.dofs_per_cell)),
symm_grad_Nx(qf_cell.size(),
std::vector<
SymmetricTensor<2, dim, NumberType>> (fe_cell.dofs_per_cell)),
grad_Nx_p_fluid(qf_cell.size(),
std::vector<
Tensor<1, dim, NumberType>>(fe_cell.dofs_per_cell))
{}
ScratchData_ASM(const ScratchData_ASM &rhs)
:
solution_total (rhs.solution_total),
fe_values_ref(rhs.fe_values_ref.get_fe(),
rhs.fe_values_ref.get_quadrature(),
rhs.fe_values_ref.get_update_flags()),
fe_face_values_ref(rhs.fe_face_values_ref.get_fe(),
rhs.fe_face_values_ref.get_quadrature(),
rhs.fe_face_values_ref.get_update_flags()),
local_dof_values(rhs.local_dof_values),
solution_grads_u_total(rhs.solution_grads_u_total),
solution_values_p_fluid_total(rhs.solution_values_p_fluid_total),
solution_grads_p_fluid_total(rhs.solution_grads_p_fluid_total),
solution_grads_face_p_fluid_total(rhs.solution_grads_face_p_fluid_total),
Nx(rhs.Nx),
Nx_p_fluid(rhs.Nx_p_fluid),
grad_Nx(rhs.grad_Nx),
symm_grad_Nx(rhs.symm_grad_Nx),
grad_Nx_p_fluid(rhs.grad_Nx_p_fluid)
{}
void reset()
{
const unsigned
int n_q_points = Nx_p_fluid.size();
const unsigned int n_dofs_per_cell = Nx_p_fluid[0].size();
Assert(local_dof_values.size() == n_dofs_per_cell, ExcInternalError());
for (unsigned int k = 0; k < n_dofs_per_cell; ++k)
{
local_dof_values[k] = 0.0;
}
Assert(solution_grads_u_total.size() == n_q_points, ExcInternalError());
Assert(solution_values_p_fluid_total.size() == n_q_points, ExcInternalError());
Assert(solution_grads_p_fluid_total.size() == n_q_points, ExcInternalError());
Assert(Nx.size() == n_q_points, ExcInternalError());
Assert(grad_Nx.size() == n_q_points, ExcInternalError());
Assert(symm_grad_Nx.size() == n_q_points, ExcInternalError());
for (unsigned int q_point = 0; q_point < n_q_points; ++q_point)
{
Assert( Nx[q_point].size() == n_dofs_per_cell, ExcInternalError());
Assert( grad_Nx[q_point].size() == n_dofs_per_cell, ExcInternalError());
Assert( symm_grad_Nx[q_point].size() == n_dofs_per_cell, ExcInternalError());
solution_grads_u_total[q_point] = 0.0;
solution_values_p_fluid_total[q_point] = 0.0;
solution_grads_p_fluid_total[q_point] = 0.0;
for (unsigned int k = 0; k < n_dofs_per_cell; ++k)
{
Nx[q_point][k] = 0.0;
Nx_p_fluid[q_point][k] = 0.0;
grad_Nx[q_point][k] = 0.0;
symm_grad_Nx[q_point][k] = 0.0;
grad_Nx_p_fluid[q_point][k] = 0.0;
}
}
const unsigned int n_f_q_points = solution_grads_face_p_fluid_total.size();
Assert(solution_grads_face_p_fluid_total.size() == n_f_q_points, ExcInternalError());
for (unsigned int f_q_point = 0; f_q_point < n_f_q_points; ++f_q_point)
solution_grads_face_p_fluid_total[f_q_point] = 0.0;
}
};
Define the boundary conditions on the mesh
template <int dim>
void Solid<dim>::make_constraints(const int &it_nr_IN)
{
pcout << " CST " << std::flush;
outfile << " CST " << std::flush;
if (it_nr_IN > 1) return;
const bool apply_dirichlet_bc = (it_nr_IN == 0);
if (apply_dirichlet_bc)
{
constraints.clear();
make_dirichlet_constraints(constraints);
}
else
{
for (unsigned int i=0; i<dof_handler_ref.n_dofs(); ++i)
if (constraints.is_inhomogeneously_constrained(i) == true)
constraints.set_inhomogeneity(i,0.0);
}
constraints.close();
}
Set-up the FE system
template <int dim>
{
timerconsole.enter_subsection("Setup system");
timerfile.enter_subsection("Setup system");
Determine number of components per block
std::vector<unsigned int> block_component(n_components, u_block);
block_component[p_fluid_component] = p_fluid_block;
The DOF handler is initialised and we renumber the grid in an efficient manner.
dof_handler_ref.distribute_dofs(fe);
void component_wise(DoFHandler< dim, spacedim > &dof_handler, const std::vector< unsigned int > &target_component=std::vector< unsigned int >())
void Cuthill_McKee(DoFHandler< dim, spacedim > &dof_handler, const bool reversed_numbering=false, const bool use_constraints=false, const std::vector< types::global_dof_index > &starting_indices=std::vector< types::global_dof_index >())
Count the number of DoFs in each block
Setup the sparsity pattern and tangent matrix
std::vector<IndexSet> all_locally_relevant_dofs
locally_owned_dofs.clear();
locally_owned_partitioning.clear();
Assert(all_locally_owned_dofs.size() > this_mpi_process, ExcInternalError());
locally_owned_dofs = all_locally_owned_dofs[this_mpi_process];
locally_relevant_dofs.clear();
locally_relevant_partitioning.clear();
Assert(all_locally_relevant_dofs.size() > this_mpi_process, ExcInternalError());
locally_relevant_dofs = all_locally_relevant_dofs[this_mpi_process];
locally_owned_partitioning.reserve(n_blocks);
locally_relevant_partitioning.reserve(n_blocks);
for (unsigned int b=0; b<n_blocks; ++b)
{
= std::accumulate(dofs_per_block.begin(),
std::next(dofs_per_block.begin(),b), 0);
= std::accumulate(dofs_per_block.begin(),
std::next(dofs_per_block.begin(),b+1), 0);
locally_owned_partitioning.push_back(locally_owned_dofs.get_view(idx_begin, idx_end));
locally_relevant_partitioning.push_back(locally_relevant_dofs.get_view(idx_begin, idx_end));
}
Print information on screen
pcout << "\nTriangulation:\n"
<< " Number of active cells: "
<< " (by partition:";
for (unsigned int p=0; p<n_mpi_processes; ++p)
pcout << (p==0 ? ' ' : '+')
pcout << ")"
<< std::endl;
pcout << " Number of degrees of freedom: "
<< dof_handler_ref.n_dofs()
<< " (by partition:";
for (unsigned int p=0; p<n_mpi_processes; ++p)
pcout << (p==0 ? ' ' : '+')
<< (
DoFTools::count_dofs_with_subdomain_association (dof_handler_ref,p));
pcout << ")"
<< std::endl;
pcout << " Number of degrees of freedom per block: "
<< "[n_u, n_p_fluid] = ["
<< dofs_per_block[u_block]
<< ", "
<< dofs_per_block[p_fluid_block]
<< "]"
<< std::endl;
unsigned int n_active_cells() const
Print information to file
outfile << "\nTriangulation:\n"
<< " Number of active cells: "
<< " (by partition:";
for (unsigned int p=0; p<n_mpi_processes; ++p)
outfile << (p==0 ? ' ' : '+')
outfile << ")"
<< std::endl;
outfile << " Number of degrees of freedom: "
<< dof_handler_ref.n_dofs()
<< " (by partition:";
for (unsigned int p=0; p<n_mpi_processes; ++p)
outfile << (p==0 ? ' ' : '+')
<< (
DoFTools::count_dofs_with_subdomain_association (dof_handler_ref,p));
outfile << ")"
<< std::endl;
outfile << " Number of degrees of freedom per block: "
<< "[n_u, n_p_fluid] = ["
<< dofs_per_block[u_block]
<< ", "
<< dofs_per_block[p_fluid_block]
<< "]"
<< std::endl;
We optimise the sparsity pattern to reflect this structure and prevent unnecessary data creation for the right-diagonal block components.
for (unsigned int ii = 0; ii < n_components; ++ii)
for (unsigned int jj = 0; jj < n_components; ++jj)
Identify "zero" matrix components of FE-system (The two components do not couple)
if (((ii == p_fluid_component) && (jj < p_fluid_component))
|| ((ii < p_fluid_component) && (jj == p_fluid_component)) )
The rest of components always couple
else
mpi_communicator);
false, this_mpi_process);
bsp.compress();
void make_sparsity_pattern(const DoFHandler< dim, spacedim > &dof_handler, SparsityPatternBase &sparsity_pattern, const AffineConstraints< number > &constraints={}, const bool keep_constrained_dofs=true, const types::subdomain_id subdomain_id=numbers::invalid_subdomain_id)
Reinitialize the (sparse) tangent matrix with the given sparsity pattern.
tangent_matrix.reinit (bsp);
Initialize the right hand side and solution vectors with number of DoFs
system_rhs.reinit(locally_owned_partitioning, mpi_communicator);
solution_n.reinit(locally_owned_partitioning, mpi_communicator);
solution_delta_OUT.
reinit(locally_owned_partitioning, mpi_communicator);
void reinit(const std::vector< IndexSet > ¶llel_partitioning, const MPI_Comm communicator=MPI_COMM_WORLD, const bool omit_zeroing_entries=false)
Non-block system
mpi_communicator);
false, this_mpi_process);
sp.compress();
tangent_matrix_nb.reinit (sp);
system_rhs_nb.reinit(locally_owned_dofs, mpi_communicator);
Set up the quadrature point history
setup_qph();
timerconsole.leave_subsection();
timerfile.leave_subsection();
}
Component extractors: used to extract sub-blocks from the global matrix Description of which local element DOFs are attached to which block component
template <int dim>
void Solid<dim>::determine_component_extractors()
{
element_indices_u.clear();
element_indices_p_fluid.clear();
for (unsigned int k = 0; k < fe.dofs_per_cell; ++k)
{
const unsigned int k_group = fe.system_to_base_index(k).first.first;
if (k_group == u_block)
element_indices_u.push_back(k);
else if (k_group == p_fluid_block)
element_indices_p_fluid.push_back(k);
else
{
Assert(k_group <= p_fluid_block, ExcInternalError());
}
}
}
Set-up quadrature point history (QPH) data objects
template <int dim>
void Solid<dim>::setup_qph()
{
pcout << "\nSetting up quadrature point data..." << std::endl;
outfile << "\nSetting up quadrature point data..." << std::endl;
Create QPH data objects.
cell_iterator end() const
active_cell_iterator begin_active(const unsigned int level=0) const
Setup the initial quadrature point data using the info stored in parameters
dof_handler_ref.begin_active()),
dof_handler_ref.end());
for (; cell!=endc; ++cell)
{
Assert(cell->is_locally_owned(), ExcInternalError());
Assert(cell->subdomain_id() == this_mpi_process, ExcInternalError());
const std::vector<std::shared_ptr<PointHistory<dim, ADNumberType> > >
lqph = quadrature_point_history.get_data(cell);
Assert(lqph.size() == n_q_points, ExcInternalError());
for (unsigned int q_point = 0; q_point < n_q_points; ++q_point)
lqph[q_point]->setup_lqp(parameters, time);
}
}
Solve the non-linear system using a Newton-Raphson scheme
Print the load step
pcout << std::endl
<< "\nTimestep "
<< time.get_timestep()
<< " @ "
<< time.get_current()
<< "s"
<< std::endl;
outfile << std::endl
<< "\nTimestep "
<< time.get_timestep()
<< " @ "
<< time.get_current()
<< "s"
<< std::endl;
Declare newton_update vector (solution of a Newton iteration), which must have as many positions as global DoFs.
(locally_owned_partitioning, mpi_communicator);
Reset the error storage objects
error_residual.reset();
error_residual_0.reset();
error_residual_norm.reset();
error_update.reset();
error_update_0.reset();
error_update_norm.reset();
print_conv_header();
Declare and initialize iterator for the Newton-Raphson algorithm steps
unsigned int newton_iteration = 0;
Iterate until error is below tolerance or max number iterations are reached
while(newton_iteration < parameters.max_iterations_NR)
{
pcout << " " << std::setw(2) << newton_iteration << " " << std::flush;
outfile << " " << std::setw(2) << newton_iteration << " " << std::flush;
Initialize global stiffness matrix and global force vector to zero
tangent_matrix = 0.0;
system_rhs = 0.0;
tangent_matrix_nb = 0.0;
system_rhs_nb = 0.0;
Apply boundary conditions
make_constraints(newton_iteration);
assemble_system(solution_delta_OUT);
Compute the rhs residual (error between external and internal forces in FE system)
get_error_residual(error_residual);
error_residual in first iteration is stored to normalize posterior error measures
if (newton_iteration == 0)
error_residual_0 = error_residual;
Determine the normalised residual error
error_residual_norm = error_residual;
error_residual_norm.normalise(error_residual_0);
If both errors are below the tolerances, exit the loop. We need to check the residual vector directly for convergence in the load steps where no external forces or displacements are imposed.
if ( ((newton_iteration > 0)
&& (error_update_norm.u <= parameters.tol_u)
&& (error_update_norm.p_fluid <= parameters.tol_p_fluid)
&& (error_residual_norm.u <= parameters.tol_f)
&& (error_residual_norm.p_fluid <= parameters.tol_f))
|| ( (newton_iteration > 0)
&& system_rhs.l2_norm() <= parameters.tol_f) )
{
pcout << "\n ***** CONVERGED! ***** "
<< system_rhs.l2_norm() << " "
<< " " << error_residual_norm.norm
<< " " << error_residual_norm.u
<< " " << error_residual_norm.p_fluid
<< " " << error_update_norm.norm
<< " " << error_update_norm.u
<< " " << error_update_norm.p_fluid
<< " " << std::endl;
outfile << "\n ***** CONVERGED! ***** "
<< system_rhs.l2_norm() << " "
<< " " << error_residual_norm.norm
<< " " << error_residual_norm.u
<< " " << error_residual_norm.p_fluid
<< " " << error_update_norm.norm
<< " " << error_update_norm.u
<< " " << error_update_norm.p_fluid
<< " " << std::endl;
print_conv_footer();
break;
}
Solve the linearized system
solve_linear_system(newton_update);
constraints.distribute(newton_update);
Compute the displacement error
get_error_update(newton_update, error_update);
error_update in first iteration is stored to normalize posterior error measures
if (newton_iteration == 0)
error_update_0 = error_update;
Determine the normalised Newton update error
error_update_norm = error_update;
error_update_norm.normalise(error_update_0);
Determine the normalised residual error
error_residual_norm = error_residual;
error_residual_norm.normalise(error_residual_0);
Print error values
pcout << " | " << std::fixed << std::setprecision(3)
<<
std::setw(7) <<
std::scientific
<< system_rhs.l2_norm()
<< " " << error_residual_norm.norm
<< " " << error_residual_norm.u
<< " " << error_residual_norm.p_fluid
<< " " << error_update_norm.norm
<< " " << error_update_norm.u
<< " " << error_update_norm.p_fluid
outfile << " | " <<
std::fixed <<
std::setprecision(3)
<<
std::setw(7) <<
std::scientific
<< system_rhs.l2_norm()
<< " " << error_residual_norm.norm
<< " " << error_residual_norm.u
<< " " << error_residual_norm.p_fluid
<< " " << error_update_norm.norm
<< " " << error_update_norm.u
<< " " << error_update_norm.p_fluid
Update
solution_delta_OUT += newton_update;
newton_update = 0.0;
newton_iteration++;
}
If maximum allowed number of iterations for Newton algorithm are reached, print non-convergence message and abort program
AssertThrow (newton_iteration < parameters.max_iterations_NR, ExcMessage(
"No convergence in nonlinear solver!"));
}
Prints the header for convergence info on console
template <int dim>
void Solid<dim>::print_conv_header()
{
static const unsigned int l_width = 120;
for (unsigned int i = 0; i < l_width; ++i)
{
pcout << "_";
outfile << "_";
}
pcout << std::endl;
outfile << std::endl;
pcout << "\n SOLVER STEP | SYS_RES "
<< "RES_NORM RES_U RES_P "
<< "NU_NORM NU_U NU_P " << std::endl;
outfile << "\n SOLVER STEP | SYS_RES "
<< "RES_NORM RES_U RES_P "
<< "NU_NORM NU_U NU_P " << std::endl;
for (unsigned int i = 0; i < l_width; ++i)
{
pcout << "_";
outfile << "_";
}
pcout << std::endl << std::endl;
outfile << std::endl << std::endl;
}
Prints the footer for convergence info on console
template <int dim>
void Solid<dim>::print_conv_footer()
{
static const unsigned int l_width = 120;
for (unsigned int i = 0; i < l_width; ++i)
{
pcout << "_";
outfile << "_";
}
pcout << std::endl << std::endl;
outfile << std::endl << std::endl;
pcout << "Relative errors:" << std::endl
<< "Displacement: "
<< error_update.u / error_update_0.u << std::endl
<< "Force (displ): "
<< error_residual.u / error_residual_0.u << std::endl
<< "Pore pressure: "
<< error_update.p_fluid / error_update_0.p_fluid << std::endl
<< "Force (pore): "
<< error_residual.p_fluid / error_residual_0.p_fluid << std::endl;
outfile << "Relative errors:" << std::endl
<< "Displacement: "
<< error_update.u / error_update_0.u << std::endl
<< "Force (displ): "
<< error_residual.u / error_residual_0.u << std::endl
<< "Pore pressure: "
<< error_update.p_fluid / error_update_0.p_fluid << std::endl
<< "Force (pore): "
<< error_residual.p_fluid / error_residual_0.p_fluid << std::endl;
}
Determine the true residual error for the problem
template <int dim>
void Solid<dim>::get_error_residual(Errors &error_residual_OUT)
{
constraints.set_zero(error_res);
error_residual_OUT.norm = error_res.l2_norm();
error_residual_OUT.u = error_res.block(u_block).l2_norm();
error_residual_OUT.p_fluid = error_res.block(p_fluid_block).l2_norm();
}
Determine the true Newton update error for the problem
template <int dim>
void Solid<dim>::get_error_update
Errors &error_update_OUT)
{
constraints.set_zero(error_ud);
error_update_OUT.norm = error_ud.l2_norm();
error_update_OUT.u = error_ud.block(u_block).l2_norm();
error_update_OUT.p_fluid = error_ud.block(p_fluid_block).l2_norm();
}
Compute the total solution, which is valid at any Newton step. This is required as, to reduce computational error, the total solution is only updated at the end of the timestep.
Cell interpolation -> Ghosted vector
solution_total (locally_owned_partitioning,
locally_relevant_partitioning,
mpi_communicator,
false);
solution_total = solution_n;
tmp = solution_delta_IN;
solution_total += tmp;
return solution_total;
}
Compute elemental stiffness tensor and right-hand side force vector, and assemble into global ones
template <int dim>
{
timerconsole.enter_subsection("Assemble system");
timerfile.enter_subsection("Assemble system");
pcout << " ASM_SYS " << std::flush;
outfile << " ASM_SYS " << std::flush;
Info given to FEValues and FEFaceValues constructors, to indicate which data will be needed at each element.
@ update_values
Shape function values.
@ update_normal_vectors
Normal vectors.
@ update_JxW_values
Transformed quadrature weights.
@ update_gradients
Shape function gradients.
@ update_quadrature_points
Transformed quadrature points.
Setup a copy of the data structures required for the process and pass them, along with the memory addresses of the assembly functions to the WorkStream object for processing
PerTaskData_ASM per_task_data(dofs_per_cell);
ScratchData_ASM<ADNumberType> scratch_data(fe, qf_cell, uf_cell,
qf_face, uf_face,
solution_total);
dof_handler_ref.begin_active()),
dof_handler_ref.end());
for (; cell != endc; ++cell)
{
Assert(cell->is_locally_owned(), ExcInternalError());
Assert(cell->subdomain_id() == this_mpi_process, ExcInternalError());
assemble_system_one_cell(cell, scratch_data, per_task_data);
copy_local_to_global_system(per_task_data);
}
timerconsole.leave_subsection();
timerfile.leave_subsection();
}
Add the local elemental contribution to the global stiffness tensor We do it twice, for the block and the non-block systems
template <int dim>
void Solid<dim>::copy_local_to_global_system (const PerTaskData_ASM &data)
{
constraints.distribute_local_to_global(data.cell_matrix,
data.cell_rhs,
data.local_dof_indices,
tangent_matrix,
system_rhs);
constraints.distribute_local_to_global(data.cell_matrix,
data.cell_rhs,
data.local_dof_indices,
tangent_matrix_nb,
system_rhs_nb);
}
Compute stiffness matrix and corresponding rhs for one element
template <int dim>
void Solid<dim>::assemble_system_one_cell
ScratchData_ASM<ADNumberType> &scratch,
PerTaskData_ASM &data) const
{
Assert(cell->is_locally_owned(), ExcInternalError());
data.reset();
scratch.reset();
scratch.fe_values_ref.reinit(cell);
cell->get_dof_indices(data.local_dof_indices);
Setup automatic differentiation
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
Initialise the dofs for the cell using the current solution.
scratch.local_dof_values[k] = scratch.solution_total[data.local_dof_indices[k]];
Mark this cell DoF as an independent variable
scratch.local_dof_values[k].diff(k, dofs_per_cell);
}
Update the quadrature point solution Compute the values and gradients of the solution in terms of the AD variables
for (unsigned int q = 0; q < n_q_points; ++q)
{
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
const unsigned int k_group = fe.system_to_base_index(k).first.first;
if (k_group == u_block)
{
scratch.fe_values_ref[u_fe].gradient(k, q);
for (unsigned int dd = 0; dd < dim; ++dd)
{
for (unsigned int ee = 0; ee < dim; ++ee)
{
scratch.solution_grads_u_total[q][dd][ee]
+= scratch.local_dof_values[k] * Grad_Nx_u[dd][ee];
}
}
}
else if (k_group == p_fluid_block)
{
const double Nx_p = scratch.fe_values_ref[p_fluid_fe].value(k, q);
scratch.fe_values_ref[p_fluid_fe].gradient(k, q);
scratch.solution_values_p_fluid_total[q]
+= scratch.local_dof_values[k] * Nx_p;
for (unsigned int dd = 0; dd < dim; ++dd)
{
scratch.solution_grads_p_fluid_total[q][dd]
+= scratch.local_dof_values[k] * Grad_Nx_p[dd];
}
}
else
Assert(k_group <= p_fluid_block, ExcInternalError());
}
}
Set up pointer "lgph" to the PointHistory object of this element
const std::vector<std::shared_ptr<const PointHistory<dim, ADNumberType> > >
lqph = quadrature_point_history.get_data(cell);
Assert(lqph.size() == n_q_points, ExcInternalError());
Precalculate the element shape function values and gradients
for (unsigned int q_point = 0; q_point < n_q_points; ++q_point)
{
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
const unsigned int i_group = fe.system_to_base_index(i).first.first;
if (i_group == u_block)
{
scratch.Nx[q_point][i] =
scratch.fe_values_ref[u_fe].value(i, q_point);
scratch.grad_Nx[q_point][i] =
scratch.fe_values_ref[u_fe].gradient(i, q_point)*F_inv_AD;
scratch.symm_grad_Nx[q_point][i] =
}
else if (i_group == p_fluid_block)
{
scratch.Nx_p_fluid[q_point][i] =
scratch.fe_values_ref[p_fluid_fe].value(i, q_point);
scratch.grad_Nx_p_fluid[q_point][i] =
scratch.fe_values_ref[p_fluid_fe].gradient(i, q_point)*F_inv_AD;
}
else
Assert(i_group <= p_fluid_block, ExcInternalError());
}
}
Assemble the stiffness matrix and rhs vector
std::vector<ADNumberType> residual_ad (dofs_per_cell, ADNumberType(0.0));
for (unsigned int q_point = 0; q_point < n_q_points; ++q_point)
{
Assert(det_F_AD > 0, ExcInternalError());
const ADNumberType p_fluid = scratch.solution_values_p_fluid_total[q_point];
{
PointHistory<dim, ADNumberType> *lqph_q_point_nc =
const_cast<PointHistory<dim, ADNumberType>*>(lqph[q_point].get());
lqph_q_point_nc->update_internal_equilibrium(F_AD);
}
Get some info from constitutive model of solid
tau_E = lqph[q_point]->get_tau_E(F_AD);
tau_fluid_vol *= -1.0 * p_fluid * det_F_AD;
Get some info from constitutive model of fluid
const ADNumberType det_F_aux = lqph[q_point]->get_converged_det_F();
= lqph[q_point]->get_overall_body_force(F_AD, parameters);
Define some aliases to make the assembly process easier to follow
const std::vector<Tensor<1,dim>> &Nu = scratch.Nx[q_point];
const std::vector<SymmetricTensor<2, dim, ADNumberType>>
&symm_grad_Nu = scratch.symm_grad_Nx[q_point];
const std::vector<double> &Np = scratch.Nx_p_fluid[q_point];
const std::vector<Tensor<1, dim, ADNumberType> > &grad_Np
= scratch.grad_Nx_p_fluid[q_point];
= scratch.solution_grads_p_fluid_total[q_point]*F_inv_AD;
const double JxW = scratch.fe_values_ref.JxW(q_point);
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
const unsigned int i_group = fe.system_to_base_index(i).first.first;
if (i_group == u_block)
{
residual_ad[i] += symm_grad_Nu[i] * ( tau_E + tau_fluid_vol ) * JxW;
residual_ad[i] -= Nu[i] * overall_body_force * JxW;
}
else if (i_group == p_fluid_block)
{
= lqph[q_point]->get_seepage_velocity_current(F_AD, grad_p);
residual_ad[i] += Np[i] * (det_F_AD - det_F_converged) * JxW;
residual_ad[i] -= time.get_delta_t() * grad_Np[i]
* seepage_vel_current * JxW;
}
else
Assert(i_group <= p_fluid_block, ExcInternalError());
}
}
Assemble the Neumann contribution (external force contribution).
for (unsigned int face = 0; face < GeometryInfo<dim>::faces_per_cell; ++face)
{
if (cell->face(face)->at_boundary() == true)
{
scratch.fe_face_values_ref.reinit(cell, face);
for (unsigned int f_q_point = 0; f_q_point < n_q_points_f; ++f_q_point)
{
= scratch.fe_face_values_ref.normal_vector(f_q_point);
= scratch.fe_face_values_ref.quadrature_point(f_q_point);
= get_neumann_traction(cell->face(face)->boundary_id(), pt, N);
const double flow
= get_prescribed_fluid_flow(cell->face(face)->boundary_id(), pt);
if ( (traction.norm() < 1e-12) && (
std::abs(flow) < 1e-12) )
continue;
const double JxW_f = scratch.fe_face_values_ref.JxW(f_q_point);
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
const unsigned int i_group = fe.system_to_base_index(i).first.first;
if ((i_group == u_block) && (traction.norm() > 1e-12))
{
const unsigned int component_i
= fe.system_to_component_index(i).first;
const double Nu_f
= scratch.fe_face_values_ref.shape_value(i, f_q_point);
residual_ad[i] -= (Nu_f * traction[component_i]) * JxW_f;
}
if ((i_group == p_fluid_block) && (
std::abs(flow) > 1e-12))
{
const double Nu_p
= scratch.fe_face_values_ref.shape_value(i, f_q_point);
residual_ad[i] -= (Nu_p * flow) * JxW_f;
}
}
}
}
}
Linearise the residual
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
const ADNumberType &R_i = residual_ad[i];
data.cell_rhs(i) -= R_i.val();
for (unsigned int j=0; j<dofs_per_cell; ++j)
data.cell_matrix(i,j) += R_i.fastAccessDx(j);
}
}
Store the converged values of the internal variables
template <int dim>
void Solid<dim>::update_end_timestep()
{
dof_handler_ref.begin_active()),
dof_handler_ref.end());
for (; cell!=endc; ++cell)
{
Assert(cell->is_locally_owned(), ExcInternalError());
Assert(cell->subdomain_id() == this_mpi_process, ExcInternalError());
const std::vector<std::shared_ptr<PointHistory<dim, ADNumberType> > >
lqph = quadrature_point_history.get_data(cell);
Assert(lqph.size() == n_q_points, ExcInternalError());
for (unsigned int q_point = 0; q_point < n_q_points; ++q_point)
lqph[q_point]->update_end_timestep();
}
}
Solve the linearized equations
template <int dim>
{
timerconsole.enter_subsection("Linear solver");
timerfile.enter_subsection("Linear solver");
pcout << " SLV " << std::flush;
outfile << " SLV " << std::flush;
newton_update_nb.
reinit(locally_owned_dofs, mpi_communicator);
1.0e-6 * system_rhs_nb.l2_norm());
solver.solve(tangent_matrix_nb, newton_update_nb, system_rhs_nb);
void reinit(const Vector &v, const bool omit_zeroing_entries=false, const bool allow_different_maps=false)
Copy the non-block solution back to block system
for (unsigned int i=0; i<locally_owned_dofs.n_elements(); ++i)
{
= locally_owned_dofs.nth_index_in_set(i);
newton_update_OUT(idx_i) = newton_update_nb(idx_i);
}
timerconsole.leave_subsection();
timerfile.leave_subsection();
}
void compress(VectorOperation::values operation)
Class to compute gradient of the pressure
template <int dim>
{
public:
GradientPostprocessor (const unsigned int p_fluid_component)
:
p_fluid_component (p_fluid_component)
{}
virtual ~GradientPostprocessor(){}
virtual void
evaluate_vector_field
{
computed_quantities.size());
for (unsigned int p=0; p<input_data.solution_gradients.size(); ++p)
{
for (unsigned int d=0; d<dim; ++d)
computed_quantities[p][d]
= input_data.solution_gradients[p][p_fluid_component][d];
}
}
private:
const unsigned int p_fluid_component;
};
#define AssertDimension(dim1, dim2)
Print results to vtu file
template <int dim> void Solid<dim>::output_results_to_vtu
(const unsigned int timestep,
const double current_time,
{
locally_relevant_partitioning,
mpi_communicator,
false);
solution_total = solution_IN;
GradientPostprocessor<dim> gradient_postprocessor(p_fluid_component);
Declare local variables with number of stress components & assign value according to "dim" value
unsigned int num_comp_symm_tensor = 6;
Declare local vectors to store values OUTPUT AVERAGED ON ELEMENTS ----------------------------------------—
std::vector<Vector<double>>cauchy_stresses_total_elements
(num_comp_symm_tensor,
std::vector<Vector<double>>cauchy_stresses_E_elements
(num_comp_symm_tensor,
std::vector<Vector<double>>stretches_elements
(dim,
std::vector<Vector<double>>seepage_velocity_elements
(dim,
OUTPUT AVERAGED ON NODES -------------------------------------------— We need to create a new FE space with a single dof per node to avoid duplication of the output on nodes for our problem with dim+1 dofs.
vertex_handler_ref.distribute_dofs(fe_vertex);
ExcDimensionMismatch(vertex_handler_ref.n_dofs(),
(vertex_handler_ref.n_dofs());
(vertex_handler_ref.n_dofs());
std::vector<Vector<double>>cauchy_stresses_total_vertex_mpi
(num_comp_symm_tensor,
std::vector<Vector<double>>sum_cauchy_stresses_total_vertex
(num_comp_symm_tensor,
std::vector<Vector<double>>cauchy_stresses_E_vertex_mpi
(num_comp_symm_tensor,
std::vector<Vector<double>>sum_cauchy_stresses_E_vertex
(num_comp_symm_tensor,
std::vector<Vector<double>>stretches_vertex_mpi
(dim,
std::vector<Vector<double>>sum_stretches_vertex
(dim,
Vector<double> porous_dissipation_vertex_mpi(vertex_handler_ref.n_dofs());
Vector<double> sum_porous_dissipation_vertex(vertex_handler_ref.n_dofs());
Vector<double> viscous_dissipation_vertex_mpi(vertex_handler_ref.n_dofs());
Vector<double> sum_viscous_dissipation_vertex(vertex_handler_ref.n_dofs());
Vector<double> solid_vol_fraction_vertex_mpi(vertex_handler_ref.n_dofs());
Vector<double> sum_solid_vol_fraction_vertex(vertex_handler_ref.n_dofs());
unsigned int n_vertices() const
We need to create a new FE space with a dim dof per node to be able to output data on nodes in vector form
vertex_vec_handler_ref.distribute_dofs(fe_vertex_vec);
ExcDimensionMismatch(vertex_vec_handler_ref.n_dofs(),
Vector<double> seepage_velocity_vertex_vec_mpi(vertex_vec_handler_ref.n_dofs());
Vector<double> sum_seepage_velocity_vertex_vec(vertex_vec_handler_ref.n_dofs());
Vector<double> counter_on_vertices_vec_mpi(vertex_vec_handler_ref.n_dofs());
Vector<double> sum_counter_on_vertices_vec(vertex_vec_handler_ref.n_dofs());
Declare and initialize local unit vectors (to construct tensor basis)
std::vector<Tensor<1,dim>> basis_vectors (dim,
Tensor<1,dim>() );
for (unsigned int i=0; i<dim; ++i)
basis_vectors[i][i] = 1;
Declare an instance of the material class object
if (parameters.mat_type == "Neo-Hooke")
NeoHooke<dim,ADNumberType> material(parameters,time);
else if (parameters.mat_type == "Ogden")
Ogden<dim,ADNumberType> material(parameters,time);
else if (parameters.mat_type == "visco-Ogden")
visco_Ogden <dim,ADNumberType>material(parameters,time);
else
Assert (
false, ExcMessage(
"Material type not implemented"));
Define a local instance of FEValues to compute updated values required to calculate stresses
Iterate through elements (cells) and Gauss Points
dof_handler_ref.begin_active()),
dof_handler_ref.end()),
vertex_handler_ref.begin_active()),
vertex_vec_handler_ref.begin_active());
start cell loop
for (; cell!=endc; ++cell, ++cell_v, ++cell_v_vec)
{
Assert(cell->is_locally_owned(), ExcInternalError());
Assert(cell->subdomain_id() == this_mpi_process, ExcInternalError());
material_id(cell->active_cell_index())=
static_cast<int>(cell->material_id());
fe_values_ref.reinit(cell);
std::vector<Tensor<2,dim>> solution_grads_u(n_q_points);
fe_values_ref[u_fe].get_function_gradients(solution_total,
solution_grads_u);
std::vector<double> solution_values_p_fluid_total(n_q_points);
fe_values_ref[p_fluid_fe].get_function_values(solution_total,
solution_values_p_fluid_total);
std::vector<Tensor<1,dim>> solution_grads_p_fluid_AD (n_q_points);
fe_values_ref[p_fluid_fe].get_function_gradients(solution_total,
solution_grads_p_fluid_AD);
start gauss point loop
for (unsigned int q_point=0; q_point<n_q_points; ++q_point)
{
const std::vector<std::shared_ptr<const PointHistory<dim,ADNumberType>>>
lqph = quadrature_point_history.get_data(cell);
Assert(lqph.size() == n_q_points, ExcInternalError());
const double p_fluid = solution_values_p_fluid_total[q_point];
Cauchy stress
lqph[q_point]->get_Cauchy_E(F_AD);
for (unsigned int i=0; i<dim; ++i)
for (unsigned int j=0; j<dim; ++j)
sigma_fluid_vol *= -p_fluid;
Volumes
const double solid_vol_fraction = (parameters.solid_vol_frac)/det_F;
Green-Lagrange strain
Seepage velocity
solution_grads_p_fluid_AD[q_point]*F_inv;
lqph[q_point]->get_seepage_velocity_current(F_AD, grad_p_fluid_AD);
Dissipations
const double porous_dissipation =
lqph[q_point]->get_porous_dissipation(F_AD, grad_p_fluid_AD);
const double viscous_dissipation =
lqph[q_point]->get_viscous_dissipation();
OUTPUT AVERAGED ON ELEMENTS ----------------------------------------— Both average on elements and on nodes is NOT weighted with the integration point volume, i.e., we assume equal contribution of each integration point to the average. Ideally, it should be weighted, but I haven't invested time in getting it to work properly.
if (parameters.outtype == "elements")
{
for (unsigned int j=0; j<dim; ++j)
{
cauchy_stresses_total_elements[j](cell->active_cell_index())
+= ((sigma*basis_vectors[j])*basis_vectors[j])/n_q_points;
cauchy_stresses_E_elements[j](cell->active_cell_index())
+= ((sigma_E*basis_vectors[j])*basis_vectors[j])/n_q_points;
stretches_elements[j](cell->active_cell_index())
/n_q_points;
seepage_velocity_elements[j](cell->active_cell_index())
}
porous_dissipation_elements(cell->active_cell_index())
+= porous_dissipation/n_q_points;
viscous_dissipation_elements(cell->active_cell_index())
+= viscous_dissipation/n_q_points;
solid_vol_fraction_elements(cell->active_cell_index())
+= solid_vol_fraction/n_q_points;
cauchy_stresses_total_elements[3](cell->active_cell_index())
+= ((sigma*basis_vectors[0])*basis_vectors[1])/n_q_points;
cauchy_stresses_total_elements[4](cell->active_cell_index())
+= ((sigma*basis_vectors[0])*basis_vectors[2])/n_q_points;
cauchy_stresses_total_elements[5](cell->active_cell_index())
+= ((sigma*basis_vectors[1])*basis_vectors[2])/n_q_points;
cauchy_stresses_E_elements[3](cell->active_cell_index())
+= ((sigma_E*basis_vectors[0])* basis_vectors[1])/n_q_points;
cauchy_stresses_E_elements[4](cell->active_cell_index())
+= ((sigma_E*basis_vectors[0])* basis_vectors[2])/n_q_points;
cauchy_stresses_E_elements[5](cell->active_cell_index())
+= ((sigma_E*basis_vectors[1])* basis_vectors[2])/n_q_points;
}
OUTPUT AVERAGED ON NODES ----------------------------------------—
else if (parameters.outtype == "nodes")
{
{
cell_v->vertex_dof_index(v, 0);
counter_on_vertices_mpi(local_vertex_indices) += 1;
for (unsigned int k=0; k<dim; ++k)
{
cauchy_stresses_total_vertex_mpi[k](local_vertex_indices)
+= (sigma*basis_vectors[k])*basis_vectors[k];
cauchy_stresses_E_vertex_mpi[k](local_vertex_indices)
+= (sigma_E*basis_vectors[k])*basis_vectors[k];
stretches_vertex_mpi[k](local_vertex_indices)
cell_v_vec->vertex_dof_index(v, k);
counter_on_vertices_vec_mpi(local_vertex_vec_indices) += 1;
seepage_velocity_vertex_vec_mpi(local_vertex_vec_indices)
}
porous_dissipation_vertex_mpi(local_vertex_indices)
+= porous_dissipation;
viscous_dissipation_vertex_mpi(local_vertex_indices)
+= viscous_dissipation;
solid_vol_fraction_vertex_mpi(local_vertex_indices)
+= solid_vol_fraction;
cauchy_stresses_total_vertex_mpi[3](local_vertex_indices)
+= (sigma*basis_vectors[0])*basis_vectors[1];
cauchy_stresses_total_vertex_mpi[4](local_vertex_indices)
+= (sigma*basis_vectors[0])*basis_vectors[2];
cauchy_stresses_total_vertex_mpi[5](local_vertex_indices)
+= (sigma*basis_vectors[1])*basis_vectors[2];
cauchy_stresses_E_vertex_mpi[3](local_vertex_indices)
+= (sigma_E*basis_vectors[0])*basis_vectors[1];
cauchy_stresses_E_vertex_mpi[4](local_vertex_indices)
+= (sigma_E*basis_vectors[0])*basis_vectors[2];
cauchy_stresses_E_vertex_mpi[5](local_vertex_indices)
+= (sigma_E*basis_vectors[1])*basis_vectors[2];
}
}
Different nodes might have different amount of contributions, e.g., corner nodes have less integration points contributing to the averaged. This is why we need a counter and divide at the end, outside the cell loop.
if (parameters.outtype == "nodes")
{
for (unsigned int d=0; d<(vertex_handler_ref.n_dofs()); ++d)
{
sum_counter_on_vertices[d] =
mpi_communicator);
sum_porous_dissipation_vertex[d] =
mpi_communicator);
sum_viscous_dissipation_vertex[d] =
mpi_communicator);
sum_solid_vol_fraction_vertex[d] =
mpi_communicator);
for (unsigned int k=0; k<num_comp_symm_tensor; ++k)
{
sum_cauchy_stresses_total_vertex[k][d] =
mpi_communicator);
sum_cauchy_stresses_E_vertex[k][d] =
mpi_communicator);
}
for (unsigned int k=0; k<dim; ++k)
{
sum_stretches_vertex[k][d] =
mpi_communicator);
}
}
for (
unsigned int d=0;
d<(vertex_vec_handler_ref.n_dofs()); ++
d)
{
sum_counter_on_vertices_vec[
d] =
mpi_communicator);
sum_seepage_velocity_vertex_vec[
d] =
mpi_communicator);
}
for (
unsigned int d=0;
d<(vertex_handler_ref.n_dofs()); ++
d)
{
if (sum_counter_on_vertices[d]>0)
{
for (unsigned int i=0; i<num_comp_symm_tensor; ++i)
{
sum_cauchy_stresses_total_vertex[i][
d] /= sum_counter_on_vertices[
d];
sum_cauchy_stresses_E_vertex[i][
d] /= sum_counter_on_vertices[
d];
}
for (unsigned int i=0; i<dim; ++i)
{
sum_stretches_vertex[i][
d] /= sum_counter_on_vertices[
d];
}
sum_porous_dissipation_vertex[
d] /= sum_counter_on_vertices[
d];
sum_viscous_dissipation_vertex[
d] /= sum_counter_on_vertices[
d];
sum_solid_vol_fraction_vertex[
d] /= sum_counter_on_vertices[
d];
}
}
for (
unsigned int d=0;
d<(vertex_vec_handler_ref.n_dofs()); ++
d)
{
if (sum_counter_on_vertices_vec[d]>0)
{
sum_seepage_velocity_vertex_vec[
d] /= sum_counter_on_vertices_vec[
d];
}
}
}
SymmetricTensor< 2, dim, Number > d(const Tensor< 2, dim, Number > &F, const Tensor< 2, dim, Number > &dF_dt)
T sum(const T &t, const MPI_Comm mpi_communicator)
Add the results to the solution to create the output file for Paraview
std::vector<DataComponentInterpretation::DataComponentInterpretation>
comp_type(dim,
std::vector<std::string> solution_name(dim, "displacement");
solution_name.push_back("pore_pressure");
data_out.attach_dof_handler(dof_handler_ref);
data_out.add_data_vector(solution_total,
solution_name,
comp_type);
data_out.add_data_vector(solution_total,
gradient_postprocessor);
partition_int.end());
data_out.add_data_vector(partitioning, "partitioning");
data_out.add_data_vector(material_id, "material_id");
@ component_is_part_of_vector
Integration point results --------------------------------------------------------—
if (parameters.outtype == "elements")
{
data_out.add_data_vector(cauchy_stresses_total_elements[0], "cauchy_xx");
data_out.add_data_vector(cauchy_stresses_total_elements[1], "cauchy_yy");
data_out.add_data_vector(cauchy_stresses_total_elements[2], "cauchy_zz");
data_out.add_data_vector(cauchy_stresses_total_elements[3], "cauchy_xy");
data_out.add_data_vector(cauchy_stresses_total_elements[4], "cauchy_xz");
data_out.add_data_vector(cauchy_stresses_total_elements[5], "cauchy_yz");
data_out.add_data_vector(cauchy_stresses_E_elements[0], "cauchy_E_xx");
data_out.add_data_vector(cauchy_stresses_E_elements[1], "cauchy_E_yy");
data_out.add_data_vector(cauchy_stresses_E_elements[2], "cauchy_E_zz");
data_out.add_data_vector(cauchy_stresses_E_elements[3], "cauchy_E_xy");
data_out.add_data_vector(cauchy_stresses_E_elements[4], "cauchy_E_xz");
data_out.add_data_vector(cauchy_stresses_E_elements[5], "cauchy_E_yz");
data_out.add_data_vector(stretches_elements[0], "stretch_xx");
data_out.add_data_vector(stretches_elements[1], "stretch_yy");
data_out.add_data_vector(stretches_elements[2], "stretch_zz");
data_out.add_data_vector(seepage_velocity_elements[0], "seepage_vel_x");
data_out.add_data_vector(seepage_velocity_elements[1], "seepage_vel_y");
data_out.add_data_vector(seepage_velocity_elements[2], "seepage_vel_z");
data_out.add_data_vector(porous_dissipation_elements, "dissipation_porous");
data_out.add_data_vector(viscous_dissipation_elements, "dissipation_viscous");
data_out.add_data_vector(solid_vol_fraction_elements, "solid_vol_fraction");
}
else if (parameters.outtype == "nodes")
{
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_total_vertex[0],
"cauchy_xx");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_total_vertex[1],
"cauchy_yy");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_total_vertex[2],
"cauchy_zz");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_total_vertex[3],
"cauchy_xy");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_total_vertex[4],
"cauchy_xz");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_total_vertex[5],
"cauchy_yz");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_E_vertex[0],
"cauchy_E_xx");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_E_vertex[1],
"cauchy_E_yy");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_E_vertex[2],
"cauchy_E_zz");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_E_vertex[3],
"cauchy_E_xy");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_E_vertex[4],
"cauchy_E_xz");
data_out.add_data_vector(vertex_handler_ref,
sum_cauchy_stresses_E_vertex[5],
"cauchy_E_yz");
data_out.add_data_vector(vertex_handler_ref,
sum_stretches_vertex[0],
"stretch_xx");
data_out.add_data_vector(vertex_handler_ref,
sum_stretches_vertex[1],
"stretch_yy");
data_out.add_data_vector(vertex_handler_ref,
sum_stretches_vertex[2],
"stretch_zz");
std::vector<DataComponentInterpretation::DataComponentInterpretation>
comp_type_vec(dim,
std::vector<std::string> solution_name_vec(dim,"seepage_velocity");
data_out.add_data_vector(vertex_vec_handler_ref,
sum_seepage_velocity_vertex_vec,
solution_name_vec,
comp_type_vec);
data_out.add_data_vector(vertex_handler_ref,
sum_porous_dissipation_vertex,
"dissipation_porous");
data_out.add_data_vector(vertex_handler_ref,
sum_viscous_dissipation_vertex,
"dissipation_viscous");
data_out.add_data_vector(vertex_handler_ref,
sum_solid_vol_fraction_vertex,
"solid_vol_fraction");
}
data_out.build_patches(degree_displ);
struct Filename
{
static std::string get_filename_vtu(unsigned int process,
unsigned int timestep,
const unsigned int n_digits = 5)
{
std::ostringstream filename_vtu;
filename_vtu
<< "solution."
<< "."
<< ".vtu";
return filename_vtu.str();
}
static std::string get_filename_pvtu(unsigned int timestep,
const unsigned int n_digits = 5)
{
std::ostringstream filename_vtu;
filename_vtu
<< "solution."
<< ".pvtu";
return filename_vtu.str();
}
static std::string get_filename_pvd (void)
{
std::ostringstream filename_vtu;
filename_vtu
<< "solution.pvd";
return filename_vtu.str();
}
};
const std::string filename_vtu = Filename::get_filename_vtu(this_mpi_process,
timestep);
std::ofstream output(filename_vtu.c_str());
data_out.write_vtu(output);
std::string int_to_string(const unsigned int value, const unsigned int digits=numbers::invalid_unsigned_int)
We have a collection of files written in parallel This next set of steps should only be performed by master process
if (this_mpi_process == 0)
{
List of all files written out at this timestep by all processors
std::vector<std::string> parallel_filenames_vtu;
for (unsigned int p=0; p<n_mpi_processes; ++p)
{
parallel_filenames_vtu.push_back(Filename::get_filename_vtu(p, timestep));
}
const std::string filename_pvtu(Filename::get_filename_pvtu(timestep));
std::ofstream pvtu_master(filename_pvtu.c_str());
data_out.write_pvtu_record(pvtu_master,
parallel_filenames_vtu);
Time dependent data master file
static std::vector<std::pair<double,std::string>> time_and_name_history;
time_and_name_history.push_back(std::make_pair(current_time,
filename_pvtu));
const std::string filename_pvd(Filename::get_filename_pvd());
std::ofstream pvd_output(filename_pvd.c_str());
}
}
void write_pvd_record(std::ostream &out, const std::vector< std::pair< double, std::string > > ×_and_names)
Print results to plotting file
template <int dim>
void Solid<dim>::output_results_to_plot(
const unsigned int timestep,
const double current_time,
std::ofstream &plotpointfile) const
{
locally_relevant_partitioning,
mpi_communicator,
false);
(void) timestep;
solution_total = solution_IN;
Variables needed to print the solution file for plotting
double total_fluid_flow = 0.0;
double total_porous_dissipation = 0.0;
double total_viscous_dissipation = 0.0;
double total_solid_vol = 0.0;
double total_vol_current = 0.0;
double total_vol_reference = 0.0;
std::vector<Point<dim+1>> solution_vertices(tracked_vertices_IN.size());
Auxiliary variables needed for mpi processing
sum_reaction_mpi = 0.0;
sum_reaction_pressure_mpi = 0.0;
sum_reaction_extra_mpi = 0.0;
double sum_total_flow_mpi = 0.0;
double sum_porous_dissipation_mpi = 0.0;
double sum_viscous_dissipation_mpi = 0.0;
double sum_solid_vol_mpi = 0.0;
double sum_vol_current_mpi = 0.0;
double sum_vol_reference_mpi = 0.0;
Declare an instance of the material class object
if (parameters.mat_type == "Neo-Hooke")
NeoHooke<dim,ADNumberType> material(parameters,time);
else if (parameters.mat_type == "Ogden")
Ogden<dim,ADNumberType> material(parameters, time);
else if (parameters.mat_type == "visco-Ogden")
visco_Ogden <dim,ADNumberType>material(parameters,time);
else
Assert (
false, ExcMessage(
"Material type not implemented"));
Define a local instance of FEValues to compute updated values required to calculate stresses
Iterate through elements (cells) and Gauss Points
dof_handler_ref.begin_active()),
dof_handler_ref.end());
start cell loop
for (; cell!=endc; ++cell)
{
Assert(cell->is_locally_owned(), ExcInternalError());
Assert(cell->subdomain_id() == this_mpi_process, ExcInternalError());
fe_values_ref.reinit(cell);
std::vector<Tensor<2,dim>> solution_grads_u(n_q_points);
fe_values_ref[u_fe].get_function_gradients(solution_total,
solution_grads_u);
std::vector<double> solution_values_p_fluid_total(n_q_points);
fe_values_ref[p_fluid_fe].get_function_values(solution_total,
solution_values_p_fluid_total);
std::vector<Tensor<1,dim >> solution_grads_p_fluid_AD(n_q_points);
fe_values_ref[p_fluid_fe].get_function_gradients(solution_total,
solution_grads_p_fluid_AD);
start gauss point loop
for (unsigned int q_point=0; q_point<n_q_points; ++q_point)
{
const std::vector<std::shared_ptr<const PointHistory<dim,ADNumberType>>>
lqph = quadrature_point_history.get_data(cell);
Assert(lqph.size() == n_q_points, ExcInternalError());
double JxW = fe_values_ref.JxW(q_point);
Volumes
sum_vol_current_mpi += det_F * JxW;
sum_vol_reference_mpi += JxW;
sum_solid_vol_mpi += parameters.solid_vol_frac * JxW * det_F;
Seepage velocity
grad_p_fluid_AD = solution_grads_p_fluid_AD[q_point]*F_inv;
= lqph[q_point]->get_seepage_velocity_current(F_AD, grad_p_fluid_AD);
Dissipations
const double porous_dissipation =
lqph[q_point]->get_porous_dissipation(F_AD, grad_p_fluid_AD);
sum_porous_dissipation_mpi += porous_dissipation * det_F * JxW;
const double viscous_dissipation = lqph[q_point]->get_viscous_dissipation();
sum_viscous_dissipation_mpi += viscous_dissipation * det_F * JxW;
Compute reaction force on load boundary & total fluid flow across drained boundary. Define a local instance of FEFaceValues to compute values required to calculate reaction force
start face loop
for (unsigned int face=0; face<GeometryInfo<dim>::faces_per_cell; ++face)
{
Reaction force
if (cell->face(face)->at_boundary() == true &&
cell->face(face)->boundary_id() == get_reaction_boundary_id_for_output() )
{
fe_face_values_ref.reinit(cell, face);
Get displacement gradients for current face
std::vector<Tensor<2,dim> > solution_grads_u_f(n_q_points_f);
fe_face_values_ref[u_fe].get_function_gradients
(solution_total,
solution_grads_u_f);
Get pressure for current element
std::vector< double > solution_values_p_fluid_total_f(n_q_points_f);
fe_face_values_ref[p_fluid_fe].get_function_values
(solution_total,
solution_values_p_fluid_total_f);
start gauss points on faces loop
for (unsigned int f_q_point=0; f_q_point<n_q_points_f; ++f_q_point)
{
const Tensor<1,dim> &N = fe_face_values_ref.normal_vector(f_q_point);
const double JxW_f = fe_face_values_ref.JxW(f_q_point);
Compute deformation gradient from displacements gradient (present configuration)
const std::vector<std::shared_ptr<const PointHistory<dim,ADNumberType>>>
lqph = quadrature_point_history.get_data(cell);
Assert(lqph.size() == n_q_points, ExcInternalError());
const double p_fluid = solution_values_p_fluid_total[f_q_point];
Cauchy stress
lqph[f_q_point]->get_Cauchy_E(F_AD);
for (unsigned int i=0; i<dim; ++i)
for (unsigned int j=0; j<dim; ++j)
sigma_fluid_vol *= -1.0*p_fluid;
sum_reaction_mpi += sigma * N * JxW_f;
sum_reaction_pressure_mpi += sigma_fluid_vol * N * JxW_f;
sum_reaction_extra_mpi += sigma_E * N * JxW_f;
}
}
Fluid flow
if (cell->face(face)->at_boundary() == true &&
(cell->face(face)->boundary_id() ==
get_drained_boundary_id_for_output().
first ||
cell->face(face)->boundary_id() ==
get_drained_boundary_id_for_output().
second ) )
{
fe_face_values_ref.reinit(cell, face);
Get displacement gradients for current face
std::vector<Tensor<2,dim>> solution_grads_u_f(n_q_points_f);
fe_face_values_ref[u_fe].get_function_gradients
(solution_total,
solution_grads_u_f);
Get pressure gradients for current face
std::vector<Tensor<1,dim>> solution_grads_p_f(n_q_points_f);
fe_face_values_ref[p_fluid_fe].get_function_gradients
(solution_total,
solution_grads_p_f);
start gauss points on faces loop
for (unsigned int f_q_point=0; f_q_point<n_q_points_f; ++f_q_point)
{
fe_face_values_ref.normal_vector(f_q_point);
const double JxW_f = fe_face_values_ref.JxW(f_q_point);
Deformation gradient and inverse from displacements gradient (present configuration)
const std::vector<std::shared_ptr<const PointHistory<dim,ADNumberType>>>
lqph = quadrature_point_history.get_data(cell);
Assert(lqph.size() == n_q_points, ExcInternalError());
Seepage velocity
= solution_grads_p_f[f_q_point]*F_inv_AD;
= lqph[f_q_point]->get_seepage_velocity_current(F_AD, grad_p);
for (unsigned int i=0; i<dim; ++i)
sum_total_flow_mpi += (seepage/det_F) * N * JxW_f;
}
}
}
}
Sum the results from different MPI process and then add to the reaction_force vector In theory, the solution on each surface (each cell) only exists in one MPI process so, we add all MPI process, one will have the solution and the others will be zero
for (unsigned int d=0; d<dim; ++d)
{
mpi_communicator);
mpi_communicator);
mpi_communicator);
}
Same for total fluid flow, and for porous and viscous dissipations
mpi_communicator);
mpi_communicator);
mpi_communicator);
mpi_communicator);
mpi_communicator);
mpi_communicator);
Extract solution for tracked vectors Copying an MPI::BlockVector into MPI::Vector is not possible, so we copy each block of MPI::BlockVector into an MPI::Vector And then we copy the MPI::Vector into "normal" Vectors
if (this_mpi_process == 0)
{
Append the pressure solution vector to the displacement solution vector, creating a single solution vector equivalent to the original BlockVector so FEFieldFunction will work with the dof_handler_ref.
+solution_u_vector.size());
for (unsigned int d=0; d<(solution_u_vector.size()); ++d)
solution_vector[d] = solution_u_vector[d];
for (unsigned int d=0; d<(solution_p_vector.size()); ++d)
solution_vector[solution_u_vector.size()+d] = solution_p_vector[d];
find_solution(dof_handler_ref, solution_vector);
for (unsigned int p=0; p<tracked_vertices_IN.size(); ++p)
{
pt_ref[0]= tracked_vertices_IN[p][0];
pt_ref[1]= tracked_vertices_IN[p][1];
pt_ref[2]= tracked_vertices_IN[p][2];
find_solution.vector_value(pt_ref, update);
for (unsigned int d=0; d<(dim+1); ++d)
{
For values close to zero, set to 0.0
if (abs(update[d])<1.5*parameters.tol_u)
update[d] = 0.0;
solution_vertices[p][d] = update[d];
}
}
Write the results to the plotting file. Add two blank lines between cycles in the cyclic loading examples so GNUPLOT can detect each cycle as a different block
if (( (parameters.geom_type == "Budday_cube_tension_compression_fully_fixed")||
(parameters.geom_type == "Budday_cube_tension_compression")||
(parameters.geom_type == "Budday_cube_shear_fully_fixed") ) &&
( (abs(current_time - parameters.end_time/3.) <0.9*parameters.delta_t)||
(abs(current_time - 2.*parameters.end_time/3.)<0.9*parameters.delta_t) ) &&
parameters.num_cycle_sets == 1 )
{
plotpointfile << std::endl<< std::endl;
}
if (( (parameters.geom_type == "Budday_cube_tension_compression_fully_fixed")||
(parameters.geom_type == "Budday_cube_tension_compression")||
(parameters.geom_type == "Budday_cube_shear_fully_fixed") ) &&
( (abs(current_time - parameters.end_time/9.) <0.9*parameters.delta_t)||
(abs(current_time - 2.*parameters.end_time/9.)<0.9*parameters.delta_t)||
(abs(current_time - 3.*parameters.end_time/9.)<0.9*parameters.delta_t)||
(abs(current_time - 5.*parameters.end_time/9.)<0.9*parameters.delta_t)||
(abs(current_time - 7.*parameters.end_time/9.)<0.9*parameters.delta_t) ) &&
parameters.num_cycle_sets == 2 )
{
plotpointfile << std::endl<< std::endl;
}
plotpointfile << std::setprecision(6) << std::scientific;
plotpointfile << std::setw(16) << current_time << ","
<< std::setw(15) << total_vol_reference << ","
<< std::setw(15) << total_vol_current << ","
<< std::setw(15) << total_solid_vol << ",";
if (current_time == 0.0)
{
for (unsigned int p=0; p<tracked_vertices_IN.size(); ++p)
{
for (
unsigned int d=0;
d<dim; ++
d)
plotpointfile << std::setw(15) << 0.0 << ",";
plotpointfile << std::setw(15) << parameters.drained_pressure << ",";
}
for (
unsigned int d=0;
d<(3*dim+2); ++
d)
plotpointfile << std::setw(15) << 0.0 << ",";
plotpointfile << std::setw(15) << 0.0;
}
else
{
for (unsigned int p=0; p<tracked_vertices_IN.size(); ++p)
for (
unsigned int d=0;
d<(dim+1); ++
d)
plotpointfile << std::setw(15) << solution_vertices[p][
d]<<
",";
for (
unsigned int d=0;
d<dim; ++
d)
plotpointfile << std::setw(15) << reaction_force[
d] <<
",";
for (
unsigned int d=0;
d<dim; ++
d)
plotpointfile << std::setw(15) << reaction_force_pressure[
d] <<
",";
for (
unsigned int d=0;
d<dim; ++
d)
plotpointfile << std::setw(15) << reaction_force_extra[
d] <<
",";
plotpointfile << std::setw(15) << total_fluid_flow << ","
<< std::setw(15) << total_porous_dissipation<< ","
<< std::setw(15) << total_viscous_dissipation;
}
plotpointfile << std::endl;
}
}
Header for console output file
template <int dim>
void Solid<dim>::print_console_file_header(std::ofstream &outputfile) const
{
outputfile << "/*-----------------------------------------------------------------------------------------";
outputfile << "\n\n Poro-viscoelastic formulation to solve nonlinear solid mechanics problems using deal.ii";
outputfile << "\n\n Problem setup by E Comellas and J-P Pelteret, University of Erlangen-Nuremberg, 2018";
outputfile << "\n\n/*-----------------------------------------------------------------------------------------";
outputfile << "\n\nCONSOLE OUTPUT: \n\n";
}
Header for plotting output file
template <int dim>
void Solid<dim>::print_plot_file_header(std::vector<
Point<dim> > &tracked_vertices,
std::ofstream &plotpointfile) const
{
plotpointfile << "#\n# *** Solution history for tracked vertices -- DOF: 0 = Ux, 1 = Uy, 2 = Uz, 3 = P ***"
<< std::endl;
for (unsigned int p=0; p<tracked_vertices.size(); ++p)
{
plotpointfile << "# Point " << p << " coordinates: ";
for (unsigned int d=0; d<dim; ++d)
{
plotpointfile << tracked_vertices[p][d];
if (!( (p == tracked_vertices.size()-1) && (d == dim-1) ))
plotpointfile << ", ";
}
plotpointfile << std::endl;
}
plotpointfile << "# The reaction force is the integral over the loaded surfaces in the "
<< "undeformed configuration of the Cauchy stress times the normal surface unit vector.\n"
<< "# reac(p) corresponds to the volumetric part of the Cauchy stress due to the pore fluid pressure"
<< " and reac(E) corresponds to the extra part of the Cauchy stress due to the solid contribution."
<< std::endl
<< "# The fluid flow is the integral over the drained surfaces in the "
<< "undeformed configuration of the seepage velocity times the normal surface unit vector."
<< std::endl
<< "# Column number:"
<< std::endl
<< "#";
unsigned int columns = 24;
for (
unsigned int d=1;
d<columns; ++
d)
plotpointfile << std::setw(15)<<
d <<
",";
plotpointfile << std::setw(15)<< columns
<< std::endl
<< "#"
<< std::right << std::setw(16) << "Time,"
<< std::right << std::setw(16) << "ref vol,"
<< std::right << std::setw(16) << "def vol,"
<< std::right << std::setw(16) << "solid vol,";
for (unsigned int p=0; p<tracked_vertices.size(); ++p)
for (
unsigned int d=0;
d<(dim+1); ++
d)
plotpointfile << std::right<< std::setw(11)
<<
"P" << p <<
"[" <<
d <<
"],";
for (
unsigned int d=0;
d<dim; ++
d)
plotpointfile << std::right<< std::setw(13)
<<
"reaction [" <<
d <<
"],";
for (
unsigned int d=0;
d<dim; ++
d)
plotpointfile << std::right<< std::setw(13)
<<
"reac(p) [" <<
d <<
"],";
for (
unsigned int d=0;
d<dim; ++
d)
plotpointfile << std::right<< std::setw(13)
<<
"reac(E) [" <<
d <<
"],";
plotpointfile << std::right<< std::setw(16)<< "fluid flow,"
<< std::right<< std::setw(16)<< "porous dissip,"
<< std::right<< std::setw(15)<< "viscous dissip"
<< std::endl;
}
Footer for console output file
template <int dim>
void Solid<dim>::print_console_file_footer(std::ofstream &outputfile) const
{
Copy "parameters" file at end of output file.
std::ifstream infile("parameters.prm");
std::string content = "";
int i;
for(i=0 ; infile.eof()!=true ; i++)
{
char aux = infile.get();
content += aux;
if(aux=='\n') content += '#';
}
i--;
content.erase(content.end()-1);
infile.close();
outputfile << "\n\n\n\n PARAMETERS FILE USED IN THIS COMPUTATION: \n#"
<< std::endl
<< content;
}
Footer for plotting output file
template <int dim>
void Solid<dim>::print_plot_file_footer(std::ofstream &plotpointfile) const
{
Copy "parameters" file at end of output file.
std::ifstream infile("parameters.prm");
std::string content = "";
int i;
for(i=0 ; infile.eof()!=true ; i++)
{
char aux = infile.get();
content += aux;
if(aux=='\n') content += '#';
}
i--;
content.erase(content.end()-1);
infile.close();
plotpointfile << "#"<< std::endl
<< "#"<< std::endl
<< "# PARAMETERS FILE USED IN THIS COMPUTATION:" << std::endl
<< "#"<< std::endl
<< content;
}
Verification examples from Ehlers and Eipper 1999
We group the definition of the geometry, boundary and loading conditions specific to the verification examples from Ehlers and Eipper 1999 into specific classes.
Base class: Tube geometry and boundary conditions
template <int dim>
class VerificationEhlers1999TubeBase
: public Solid<dim>
{
public:
VerificationEhlers1999TubeBase (const Parameters::AllParameters ¶meters)
: Solid<dim> (parameters)
{}
virtual ~VerificationEhlers1999TubeBase () {}
private:
virtual void make_grid() override
{
0.1,
0.5);
this->triangulation.reset_manifold(0);
this->triangulation.set_manifold (0, manifold_description_3d);
this->triangulation.refine_global(
std::max (1U, this->parameters.global_refinement));
this->triangulation.reset_manifold(0);
}
virtual void define_tracked_vertices(std::vector<
Point<dim> > &tracked_vertices)
override
{
tracked_vertices[0][0] = 0.0*this->parameters.scale;
tracked_vertices[0][1] = 0.0*this->parameters.scale;
tracked_vertices[0][2] = 0.5*this->parameters.scale;
tracked_vertices[1][0] = 0.0*this->parameters.scale;
tracked_vertices[1][1] = 0.0*this->parameters.scale;
tracked_vertices[1][2] = -0.5*this->parameters.scale;
}
{
if (this->time.get_timestep() < 2)
{
2,
constraints,
(this->fe.component_mask(this->pressure)));
}
else
{
2,
constraints,
(this->fe.component_mask(this->pressure)));
}
0,
constraints,
(this->fe.component_mask(this->x_displacement)|
this->fe.component_mask(this->y_displacement) ) );
1,
constraints,
(this->fe.component_mask(this->x_displacement) |
this->fe.component_mask(this->y_displacement) |
this->fe.component_mask(this->z_displacement) ));
}
virtual double
{
(void)pt;
return 0.0;
}
get_reaction_boundary_id_for_output() const override
{
return 2;
}
virtual std::pair<types::boundary_id,types::boundary_id>
get_drained_boundary_id_for_output() const override
{
return std::make_pair(2,2);
}
virtual std::vector<double>
const int &direction) const override
{
std::vector<double> displ_incr(dim, 0.0);
(void)direction;
AssertThrow(
false, ExcMessage(
"Displacement loading not implemented for Ehlers verification examples."));
return displ_incr;
}
};
void cylinder(Triangulation< dim > &tria, const double radius=1., const double half_length=1.)
static constexpr double PI
::VectorizedArray< Number, width > max(const ::VectorizedArray< Number, width > &, const ::VectorizedArray< Number, width > &)
Derived class: Step load example
template <int dim>
class VerificationEhlers1999StepLoad
: public VerificationEhlers1999TubeBase<dim>
{
public:
VerificationEhlers1999StepLoad (const Parameters::AllParameters ¶meters)
: VerificationEhlers1999TubeBase<dim> (parameters)
{}
virtual ~VerificationEhlers1999StepLoad () {}
private:
{
if (this->parameters.load_type == "pressure")
{
if (boundary_id == 2)
{
return this->parameters.load * N;
}
}
(void)pt;
}
};
Derived class: Load increasing example
template <int dim>
class VerificationEhlers1999IncreaseLoad
: public VerificationEhlers1999TubeBase<dim>
{
public:
VerificationEhlers1999IncreaseLoad (const Parameters::AllParameters ¶meters)
: VerificationEhlers1999TubeBase<dim> (parameters)
{}
virtual ~VerificationEhlers1999IncreaseLoad () {}
private:
{
if (this->parameters.load_type == "pressure")
{
if (boundary_id == 2)
{
const double initial_load = this->parameters.load;
const double final_load = 20.0*initial_load;
const double initial_time = this->time.get_delta_t();
const double final_time = this->time.get_end();
const double current_time = this->time.get_current();
const double load = initial_load + (final_load-initial_load)*(current_time-initial_time)/(final_time-initial_time);
return load * N;
}
}
(void)pt;
}
};
Class: Consolidation cube
template <int dim>
class VerificationEhlers1999CubeConsolidation
: public Solid<dim>
{
public:
VerificationEhlers1999CubeConsolidation (const Parameters::AllParameters ¶meters)
: Solid<dim> (parameters)
{}
virtual ~VerificationEhlers1999CubeConsolidation () {}
private:
virtual void
make_grid() override
{
true);
this->triangulation.refine_global(
std::max (1U, this->parameters.global_refinement));
this->triangulation.begin_active(), endc = this->triangulation.end();
for (; cell != endc; ++cell)
{
for (unsigned int face = 0; face < GeometryInfo<dim>::faces_per_cell; ++face)
if (cell->face(face)->at_boundary() == true &&
cell->face(face)->center()[2] == 1.0 * this->parameters.scale)
{
if (cell->face(face)->center()[0] < 0.5 * this->parameters.scale &&
cell->face(face)->center()[1] < 0.5 * this->parameters.scale)
cell->face(face)->set_boundary_id(100);
else
cell->face(face)->set_boundary_id(101);
}
}
}
virtual void
define_tracked_vertices(std::vector<
Point<dim> > &tracked_vertices)
override {
tracked_vertices[0][0] = 0.0*this->parameters.scale;
tracked_vertices[0][1] = 0.0*this->parameters.scale;
tracked_vertices[0][2] = 1.0*this->parameters.scale;
tracked_vertices[1][0] = 0.0*this->parameters.scale;
tracked_vertices[1][1] = 0.0*this->parameters.scale;
tracked_vertices[1][2] = 0.0*this->parameters.scale;
}
virtual void
{
if (this->time.get_timestep() < 2)
{
101,
constraints,
(this->fe.component_mask(this->pressure)));
}
else
{
101,
constraints,
(this->fe.component_mask(this->pressure)));
}
0,
constraints,
this->fe.component_mask(this->x_displacement));
1,
constraints,
this->fe.component_mask(this->x_displacement));
2,
constraints,
this->fe.component_mask(this->y_displacement));
3,
constraints,
this->fe.component_mask(this->y_displacement));
4,
constraints,
( this->fe.component_mask(this->x_displacement) |
this->fe.component_mask(this->y_displacement) |
this->fe.component_mask(this->z_displacement) ));
}
{
if (this->parameters.load_type == "pressure")
{
if (boundary_id == 100)
{
return this->parameters.load * N;
}
}
(void)pt;
}
virtual double
{
(void)pt;
return 0.0;
}
get_reaction_boundary_id_for_output() const override
{
return 100;
}
virtual std::pair<types::boundary_id,types::boundary_id>
get_drained_boundary_id_for_output() const override
{
return std::make_pair(101,101);
}
virtual std::vector<double>
const int &direction) const override
{
std::vector<double> displ_incr(dim, 0.0);
(void)direction;
AssertThrow(
false, ExcMessage(
"Displacement loading not implemented for Ehlers verification examples."));
return displ_incr;
}
};
void hyper_rectangle(Triangulation< dim, spacedim > &tria, const Point< dim > &p1, const Point< dim > &p2, const bool colorize=false)
Franceschini experiments
template <int dim>
class Franceschini2006Consolidation
: public Solid<dim>
{
public:
Franceschini2006Consolidation (const Parameters::AllParameters ¶meters)
: Solid<dim> (parameters)
{}
virtual ~Franceschini2006Consolidation () {}
private:
virtual void make_grid() override
{
const Point<dim-1> mesh_center(0.0, 0.0);
const double radius = 0.5;
const double height = 0.27; //8.1 mm for 30 mm radius
const double height = 0.23;
mesh_center,
radius);
2,
height,
this->triangulation);
this->triangulation.set_manifold(cylinder_id, cylinder_3d);
{
for (unsigned
int face = 0; face < GeometryInfo<dim>::faces_per_cell; ++face)
{
if (cell->face(face)->at_boundary() == true)
{
if (cell->face(face)->center()[2] == 0.0)
cell->face(face)->set_boundary_id(1);
else if (cell->face(face)->center()[2] == height)
cell->face(face)->set_boundary_id(2);
else
{
cell->face(face)->set_boundary_id(0);
cell->face(face)->set_all_manifold_ids(cylinder_id);
}
}
}
}
this->triangulation.refine_global(
std::max (1U, this->parameters.global_refinement));
}
virtual void define_tracked_vertices(std::vector<
Point<dim> > &tracked_vertices)
override
{
tracked_vertices[0][0] = 0.0*this->parameters.scale;
tracked_vertices[0][1] = 0.0*this->parameters.scale;
void hyper_ball(Triangulation< dim > &tria, const Point< dim > ¢er=Point< dim >(), const double radius=1., const bool attach_spherical_manifold_on_boundary_cells=false)
void extrude_triangulation(const Triangulation< 2, 2 > &input, const unsigned int n_slices, const double height, Triangulation< 3, 3 > &result, const bool copy_manifold_ids=false, const std::vector< types::manifold_id > &manifold_priorities={})
tracked_vertices[0][2] = 0.27*this->parameters.scale;
tracked_vertices[0][2] = 0.23*this->parameters.scale;
tracked_vertices[1][0] = 0.0*this->parameters.scale;
tracked_vertices[1][1] = 0.0*this->parameters.scale;
tracked_vertices[1][2] = 0.0*this->parameters.scale;
}
{
if (this->time.get_timestep() < 2)
{
1,
constraints,
(this->fe.component_mask(this->pressure)));
2,
constraints,
(this->fe.component_mask(this->pressure)));
}
else
{
1,
constraints,
(this->fe.component_mask(this->pressure)));
2,
constraints,
(this->fe.component_mask(this->pressure)));
}
0,
constraints,
(this->fe.component_mask(this->x_displacement)|
this->fe.component_mask(this->y_displacement) ) );
1,
constraints,
(this->fe.component_mask(this->x_displacement) |
this->fe.component_mask(this->y_displacement) |
this->fe.component_mask(this->z_displacement) ));
2,
constraints,
(this->fe.component_mask(this->x_displacement) |
this->fe.component_mask(this->y_displacement) ));
}
virtual double
{
(void)pt;
return 0.0;
}
get_reaction_boundary_id_for_output() const override
{
return 2;
}
virtual std::pair<types::boundary_id,types::boundary_id>
get_drained_boundary_id_for_output() const override
{
return std::make_pair(1,2);
}
virtual std::vector<double>
const int &direction) const override
{
std::vector<double> displ_incr(dim, 0.0);
(void)direction;
AssertThrow(
false, ExcMessage(
"Displacement loading not implemented for Franceschini examples."));
return displ_incr;
}
{
if (this->parameters.load_type == "pressure")
{
if (boundary_id == 2)
{
return (this->parameters.load * N);
}
}
(void)pt;
}
};
Examples to reproduce experiments by Budday et al. 2017
We group the definition of the geometry, boundary and loading conditions specific to the examples to reproduce experiments by Budday et al. 2017 into specific classes.
Base class: Cube geometry and loading pattern
template <int dim>
class BrainBudday2017BaseCube
: public Solid<dim>
{
public:
BrainBudday2017BaseCube (const Parameters::AllParameters ¶meters)
: Solid<dim> (parameters)
{}
virtual ~BrainBudday2017BaseCube () {}
private:
virtual void
make_grid() override
{
0.0,
1.0,
true);
this->triangulation.begin_active(), endc = this->triangulation.end();
for (; cell != endc; ++cell)
{
for (unsigned int face = 0; face < GeometryInfo<dim>::faces_per_cell; ++face)
if (cell->face(face)->at_boundary() == true &&
( cell->face(face)->boundary_id() == 0 ||
cell->face(face)->boundary_id() == 1 ||
cell->face(face)->boundary_id() == 2 ||
cell->face(face)->boundary_id() == 3 ) )
cell->face(face)->set_boundary_id(100);
}
this->triangulation.refine_global(
std::max (1U, this->parameters.global_refinement));
}
virtual double
{
(void)pt;
return 0.0;
}
virtual std::pair<types::boundary_id,types::boundary_id>
get_drained_boundary_id_for_output() const override
{
return std::make_pair(100,100);
}
};
void hyper_cube(Triangulation< dim, spacedim > &tria, const double left=0., const double right=1., const bool colorize=false)
Derived class: Uniaxial boundary conditions
template <int dim>
class BrainBudday2017CubeTensionCompression
: public BrainBudday2017BaseCube<dim>
{
public:
BrainBudday2017CubeTensionCompression (const Parameters::AllParameters ¶meters)
: BrainBudday2017BaseCube<dim> (parameters)
{}
virtual ~BrainBudday2017CubeTensionCompression () {}
private:
virtual void
define_tracked_vertices(std::vector<
Point<dim> > &tracked_vertices)
override {
tracked_vertices[0][0] = 0.5*this->parameters.scale;
tracked_vertices[0][1] = 0.5*this->parameters.scale;
tracked_vertices[0][2] = 1.0*this->parameters.scale;
tracked_vertices[1][0] = 0.5*this->parameters.scale;
tracked_vertices[1][1] = 0.5*this->parameters.scale;
tracked_vertices[1][2] = 0.5*this->parameters.scale;
}
virtual void
{
if (this->time.get_timestep() < 2)
{
100,
constraints,
(this->fe.component_mask(this->pressure)));
}
else
{
100,
constraints,
(this->fe.component_mask(this->pressure)));
}
4,
constraints,
this->fe.component_mask(this->z_displacement) );
Point<dim> fix_node(0.5*this->parameters.scale, 0.5*this->parameters.scale, 0.0);
cell = this->dof_handler_ref.begin_active(), endc = this->dof_handler_ref.end();
for (; cell != endc; ++cell)
for (unsigned int node = 0; node < GeometryInfo<dim>::vertices_per_cell; ++node)
{
if ( (
abs(cell->vertex(node)[2]-fix_node[2]) < (1e-6 * this->parameters.scale))
&& (
abs(cell->vertex(node)[0]-fix_node[0]) < (1e-6 * this->parameters.scale)))
constraints.add_line(cell->vertex_dof_index(node, 0));
if ( (
abs(cell->vertex(node)[2]-fix_node[2]) < (1e-6 * this->parameters.scale))
&& (
abs(cell->vertex(node)[1]-fix_node[1]) < (1e-6 * this->parameters.scale)))
constraints.add_line(cell->vertex_dof_index(node, 1));
}
if (this->parameters.load_type == "displacement")
{
const std::vector<double>
value = get_dirichlet_load(5,2);
5,
constraints,
this->fe.component_mask(direction));
}
}
{
if (this->parameters.load_type == "pressure")
{
if (boundary_id == 5)
{
const double final_load = this->parameters.load;
const double current_time = this->time.get_current();
const double final_time = this->time.get_end();
const double num_cycles = 3.0;
return final_load/2.0 * (1.0 -
std::sin(
numbers::PI * (2.0*num_cycles*current_time/final_time + 0.5))) * N;
}
}
(void)pt;
}
get_reaction_boundary_id_for_output() const override
{
return 5;
}
virtual std::vector<double>
const int &direction) const override
{
std::vector<double> displ_incr(dim,0.0);
if ( (boundary_id == 5) && (direction == 2) )
{
const double final_displ = this->parameters.load;
const double current_time = this->time.get_current();
const double final_time = this->time.get_end();
const double delta_time = this->time.get_delta_t();
const double num_cycles = 3.0;
double current_displ = 0.0;
double previous_displ = 0.0;
if (this->parameters.num_cycle_sets == 1)
{
current_displ = final_displ/2.0 * (1.0
previous_displ = final_displ/2.0 * (1.0
}
else
{
if ( current_time <= (final_time*1.0/3.0) )
{
(2.0*num_cycles*current_time/(final_time*1.0/3.0) + 0.5)));
(2.0*num_cycles*(current_time-delta_time)/(final_time*1.0/3.0) + 0.5)));
}
else
{
(2.0*num_cycles*current_time / (final_time*2.0/3.0)
- (num_cycles - 0.5) )));
(2.0*num_cycles*(current_time-delta_time) / (final_time*2.0/3.0)
- (num_cycles - 0.5))));
}
}
displ_incr[2] = current_displ - previous_displ;
}
return displ_incr;
}
};
::VectorizedArray< Number, width > sin(const ::VectorizedArray< Number, width > &)
Derived class: No lateral displacement in loading surfaces
template <int dim>
class BrainBudday2017CubeTensionCompressionFullyFixed
: public BrainBudday2017BaseCube<dim>
{
public:
BrainBudday2017CubeTensionCompressionFullyFixed (const Parameters::AllParameters ¶meters)
: BrainBudday2017BaseCube<dim> (parameters)
{}
virtual ~BrainBudday2017CubeTensionCompressionFullyFixed () {}
private:
virtual void
define_tracked_vertices(std::vector<
Point<dim> > &tracked_vertices)
override {
tracked_vertices[0][0] = 0.5*this->parameters.scale;
tracked_vertices[0][1] = 0.5*this->parameters.scale;
tracked_vertices[0][2] = 1.0*this->parameters.scale;
tracked_vertices[1][0] = 0.5*this->parameters.scale;
tracked_vertices[1][1] = 0.5*this->parameters.scale;
tracked_vertices[1][2] = 0.5*this->parameters.scale;
}
virtual void
{
if (this->time.get_timestep() < 2)
{
100,
constraints,
(this->fe.component_mask(this->pressure)));
}
else
{
100,
constraints,
(this->fe.component_mask(this->pressure)));
}
4,
constraints,
(this->fe.component_mask(this->x_displacement) |
this->fe.component_mask(this->y_displacement) |
this->fe.component_mask(this->z_displacement) ));
if (this->parameters.load_type == "displacement")
{
const std::vector<double>
value = get_dirichlet_load(5,2);
5,
constraints,
this->fe.component_mask(direction) );
5,
constraints,
(this->fe.component_mask(this->x_displacement) |
this->fe.component_mask(this->y_displacement) ));
}
}
{
if (this->parameters.load_type == "pressure")
{
if (boundary_id == 5)
{
const double final_load = this->parameters.load;
const double current_time = this->time.get_current();
const double final_time = this->time.get_end();
const double num_cycles = 3.0;
return final_load/2.0 * (1.0 -
std::sin(
numbers::PI * (2.0*num_cycles*current_time/final_time + 0.5))) * N;
}
}
(void)pt;
}
get_reaction_boundary_id_for_output() const override
{
return 5;
}
virtual std::vector<double>
const int &direction) const override
{
std::vector<double> displ_incr(dim,0.0);
if ( (boundary_id == 5) && (direction == 2) )
{
const double final_displ = this->parameters.load;
const double current_time = this->time.get_current();
const double final_time = this->time.get_end();
const double delta_time = this->time.get_delta_t();
const double num_cycles = 3.0;
double current_displ = 0.0;
double previous_displ = 0.0;
if (this->parameters.num_cycle_sets == 1)
{
current_displ = final_displ/2.0 * (1.0 -
std::sin(
numbers::PI * (2.0*num_cycles*current_time/final_time + 0.5)));
previous_displ = final_displ/2.0 * (1.0 -
std::sin(
numbers::PI * (2.0*num_cycles*(current_time-delta_time)/final_time + 0.5)));
}
else
{
if ( current_time <= (final_time*1.0/3.0) )
{
(2.0*num_cycles*current_time/(final_time*1.0/3.0) + 0.5)));
(2.0*num_cycles*(current_time-delta_time)/(final_time*1.0/3.0) + 0.5)));
}
else
{
(2.0*num_cycles*current_time / (final_time*2.0/3.0)
- (num_cycles - 0.5) )));
(2.0*num_cycles*(current_time-delta_time) / (final_time*2.0/3.0)
- (num_cycles - 0.5))));
}
}
displ_incr[2] = current_displ - previous_displ;
}
return displ_incr;
}
};
Derived class: No lateral or vertical displacement in loading surface
template <int dim>
class BrainBudday2017CubeShearFullyFixed
: public BrainBudday2017BaseCube<dim>
{
public:
BrainBudday2017CubeShearFullyFixed (const Parameters::AllParameters ¶meters)
: BrainBudday2017BaseCube<dim> (parameters)
{}
virtual ~BrainBudday2017CubeShearFullyFixed () {}
private:
virtual void
define_tracked_vertices(std::vector<
Point<dim> > &tracked_vertices)
override {
tracked_vertices[0][0] = 0.75*this->parameters.scale;
tracked_vertices[0][1] = 0.5*this->parameters.scale;
tracked_vertices[0][2] = 0.0*this->parameters.scale;
tracked_vertices[1][0] = 0.25*this->parameters.scale;
tracked_vertices[1][1] = 0.5*this->parameters.scale;
tracked_vertices[1][2] = 0.0*this->parameters.scale;
}
virtual void
{
if (this->time.get_timestep() < 2)
{
100,
constraints,
(this->fe.component_mask(this->pressure)));
}
else
{
100,
constraints,
(this->fe.component_mask(this->pressure)));
}
5,
constraints,
(this->fe.component_mask(this->x_displacement) |
this->fe.component_mask(this->y_displacement) |
this->fe.component_mask(this->z_displacement) ));
if (this->parameters.load_type == "displacement")
{
const std::vector<double>
value = get_dirichlet_load(4,0);
4,
constraints,
this->fe.component_mask(direction));
4,
constraints,
(this->fe.component_mask(this->y_displacement) |
this->fe.component_mask(this->z_displacement) ));
}
}
{
if (this->parameters.load_type == "pressure")
{
if (boundary_id == 4)
{
const double final_load = this->parameters.load;
const double current_time = this->time.get_current();
const double final_time = this->time.get_end();
const double num_cycles = 3.0;
return (final_load * (
std::sin(2.0*(
numbers::PI)*num_cycles*current_time/final_time)) * (R * N));
}
}
(void)pt;
}
get_reaction_boundary_id_for_output() const override
{
return 4;
}
virtual std::vector<double>
const int &direction) const override
{
std::vector<double> displ_incr (dim, 0.0);
if ( (boundary_id == 4) && (direction == 0) )
{
const double final_displ = this->parameters.load;
const double current_time = this->time.get_current();
const double final_time = this->time.get_end();
const double delta_time = this->time.get_delta_t();
const double num_cycles = 3.0;
double current_displ = 0.0;
double previous_displ = 0.0;
if (this->parameters.num_cycle_sets == 1)
{
current_displ = final_displ * (
std::sin(2.0*(
numbers::PI)*num_cycles*current_time/final_time));
previous_displ = final_displ * (
std::sin(2.0*(
numbers::PI)*num_cycles*(current_time-delta_time)/final_time));
}
else
{
AssertThrow(
false, ExcMessage(
"Problem type not defined. Budday shear experiments implemented only for one set of cycles."));
}
displ_incr[0] = current_displ - previous_displ;
}
return displ_incr;
}
};
}
Number angle(const Tensor< 1, spacedim, Number > &a, const Tensor< 1, spacedim, Number > &b)
Main function
Lastly we provide the main driver function which is similar to the other tutorials.
int main (int argc, char *argv[])
{
using namespace NonLinearPoroViscoElasticity;
const unsigned int n_tbb_processes = 1;
try
{
Parameters::AllParameters parameters ("parameters.prm");
if (parameters.geom_type == "Ehlers_tube_step_load")
{
VerificationEhlers1999StepLoad<3> solid_3d(parameters);
solid_3d.run();
}
else if (parameters.geom_type == "Ehlers_tube_increase_load")
{
VerificationEhlers1999IncreaseLoad<3> solid_3d(parameters);
solid_3d.run();
}
else if (parameters.geom_type == "Ehlers_cube_consolidation")
{
VerificationEhlers1999CubeConsolidation<3> solid_3d(parameters);
solid_3d.run();
}
else if (parameters.geom_type == "Franceschini_consolidation")
{
Franceschini2006Consolidation<3> solid_3d(parameters);
solid_3d.run();
}
else if (parameters.geom_type == "Budday_cube_tension_compression")
{
BrainBudday2017CubeTensionCompression<3> solid_3d(parameters);
solid_3d.run();
}
else if (parameters.geom_type == "Budday_cube_tension_compression_fully_fixed")
{
BrainBudday2017CubeTensionCompressionFullyFixed<3> solid_3d(parameters);
solid_3d.run();
}
else if (parameters.geom_type == "Budday_cube_shear_fully_fixed")
{
BrainBudday2017CubeShearFullyFixed<3> solid_3d(parameters);
solid_3d.run();
}
else
{
AssertThrow(
false, ExcMessage(
"Problem type not defined. Current setting: " + parameters.geom_type));
}
}
catch (std::exception &exc)
{
{
std::cerr << std::endl << std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Exception on processing: " << std::endl << exc.what()
<< std::endl << "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
}
catch (...)
{
{
std::cerr << std::endl << std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Unknown exception!" << std::endl << "Aborting!"
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
}
return 0;
}
unsigned int this_mpi_process(const MPI_Comm mpi_communicator)
























