 |
Reference documentation for deal.II version 9.6.0
|
 |
Reference documentation for deal.II version 9.6.0
|
This tutorial depends on step-6.
In this example, our aims are the following:
While the second aim is difficult to describe in general terms without reference to the code, we will discuss the other two aims in the following. The use of multiple threads will then be detailed at the relevant places within the program. We will, however, follow the general discussion of the WorkStream approach detailed in the Parallel computing with multiple processors accessing shared memory documentation topic.
In the present example program, we want to numerically approximate the solution of the advection equation
\beta \cdot \nabla u = f,
where \beta is a vector field that describes the advection direction and speed (which may be dependent on the space variables if \beta=\beta(\mathbf x)), f is a source function, and u is the solution. The physical process that this equation describes is that of a given flow field \beta, with which another substance is transported, the density or concentration of which is given by u. The equation does not contain diffusion of this second species within its carrier substance, but there are source terms.
It is obvious that at the inflow, the above equation needs to be augmented by boundary conditions:
u = g \qquad\qquad \mathrm{on}\ \partial\Omega_-,
where \partial\Omega_- describes the inflow portion of the boundary and is formally defined by
\partial\Omega_- = \{{\mathbf x}\in \partial\Omega: \beta\cdot{\mathbf n}({\mathbf x}) < 0\},
and {\mathbf n}({\mathbf x}) being the outward normal to the domain at point {\mathbf x}\in\partial\Omega. This definition is quite intuitive, since as {\mathbf n} points outward, the scalar product with \beta can only be negative if the transport direction \beta points inward, i.e. at the inflow boundary. The mathematical theory states that we must not pose any boundary condition on the outflow part of the boundary.
Unfortunately, the equation stated above cannot be solved in a stable way using the standard finite element method. The problem is that solutions to this equation possess insufficient regularity perpendicular to the transport direction: while they are smooth along the streamlines defined by the "wind field" \beta, they may be discontinuous perpendicular to this direction. This is easy to understand: what the equation \beta \cdot \nabla u = f means is in essence that the rate of change of u in direction \beta equals f. But the equation has no implications for the derivatives in the perpendicular direction, and consequently if u is discontinuous at a point on the inflow boundary, then this discontinuity will simply be transported along the streamline of the wind field that starts at this boundary point. These discontinuities lead to numerical instabilities that make a stable solution by a standard continuous finite element discretization impossible.
A standard approach to address this difficulty is the "streamline-upwind Petrov-Galerkin" (SUPG) method, sometimes also called the streamline diffusion method. A good explanation of the method can be found in [79] . Formally, this method replaces the step in which we derive the weak form of the differential equation from the strong form: Instead of multiplying the equation by a test function v and integrating over the domain, we instead multiply by v + \delta \beta\cdot\nabla v, where \delta is a parameter that is chosen in the range of the (local) mesh width h; good results are usually obtained by setting \delta=0.1h. (Why this is called "streamline diffusion" will be explained below; for the moment, let us simply take for granted that this is how we derive a stable discrete formulation.) The value for \delta here is small enough that we do not introduce excessive diffusion, but large enough that the resulting problem is well-posed.
Using the test functions as defined above, an initial weak form of the problem would ask for finding a function u_h so that for all test functions v_h we have
(\beta \cdot \nabla u_h, v_h + \delta \beta\cdot\nabla v_h)_\Omega = (f, v_h + \delta \beta\cdot\nabla v_h)_\Omega.
However, we would like to include inflow boundary conditions u=g weakly into this problem, and this can be done by requiring that in addition to the equation above we also have
(u_h, w_h)_{\partial\Omega_-} = (g, w_h)_{\partial\Omega_-}
for all test functions w_h that live on the boundary and that are from a suitable test space. It turns out that a suitable space of test functions happens to be \beta\cdot {\mathbf n} times the traces of the functions v_h in the test space we already use for the differential equation in the domain. Thus, we require that for all test functions v_h we have
(u_h, \beta\cdot {\mathbf n} v_h)_{\partial\Omega_-} = (g, \beta\cdot {\mathbf n} v_h)_{\partial\Omega_-}.
Without attempting a justification (see again the literature on the finite element method in general, and the streamline diffusion method in particular), we can combine the equations for the differential equation and the boundary values in the following weak formulation of our stabilized problem: find a discrete function u_h such that for all discrete test functions v_h there holds
(\beta \cdot \nabla u_h, v_h + \delta \beta\cdot\nabla v_h)_\Omega - (u_h, \beta\cdot {\mathbf n} v_h)_{\partial\Omega_-} = (f, v_h + \delta \beta\cdot\nabla v_h)_\Omega - (g, \beta\cdot {\mathbf n} v_h)_{\partial\Omega_-}.
One would think that this leads to a system matrix to be inverted of the form
a_{ij} = (\beta \cdot \nabla \varphi_i, \varphi_j + \delta \beta\cdot\nabla \varphi_j)_\Omega - (\varphi_i, \beta\cdot {\mathbf n} \varphi_j)_{\partial\Omega_-},
with basis functions \varphi_i,\varphi_j. However, this is a pitfall that happens to every numerical analyst at least once (including the author): we have here expanded the solution u_h = \sum_i U_i \varphi_i, but if we do so, we will have to solve the problem
U^T A = F^T,
where U is the vector of expansion coefficients, i.e., we have to solve the transpose problem of what we might have expected naively.
This is a point we made in the introduction of step-3. There, we argued that to avoid this very kind of problem, one should get in the habit of always multiplying with test functions from the left instead of from the right to obtain the correct matrix right away. In order to obtain the form of the linear system that we need, it is therefore best to rewrite the weak formulation to
(v_h + \delta \beta\cdot\nabla v_h, \beta \cdot \nabla u_h)_\Omega - (\beta\cdot {\mathbf n} v_h, u_h)_{\partial\Omega_-} = (v_h + \delta \beta\cdot\nabla v_h, f)_\Omega - (\beta\cdot {\mathbf n} v_h, g)_{\partial\Omega_-}
and then to obtain
a_{ij} = (\varphi_i + \delta \beta \cdot \nabla \varphi_i, \beta\cdot\nabla \varphi_j)_\Omega - (\beta\cdot {\mathbf n} \varphi_i, \varphi_j)_{\partial\Omega_-},
as system matrix. We will assemble this matrix in the program.
Looking at the bilinear form mentioned above, we see that the discrete solution has to satisfy an equation of which the left hand side in weak form has a domain term of the kind
(v_h + \delta \beta\cdot\nabla v_h, \beta \cdot \nabla u_h)_\Omega,
or if we split this up, the form
(v_h, \beta \cdot \nabla u_h)_\Omega + (\delta \beta\cdot\nabla v_h, \beta \cdot \nabla u_h)_\Omega.
If we wanted to see what strong form of the equation that would correspond to, we need to integrate the second term. This yields the following formulation, where for simplicity we'll ignore boundary terms for now:
(v_h, \beta \cdot \nabla u_h)_\Omega - \left(v_h, \delta \nabla \cdot \left[\beta \left(\beta \cdot \nabla u_h\right)\right]\right)_\Omega + \text{boundary terms}.
Let us assume for a moment that the wind field \beta is divergence-free, i.e., that \nabla \cdot \beta = 0. Then applying the product rule to the derivative of the term in square brackets on the right and using the divergence-freeness will give us the following:
(v_h, \beta \cdot \nabla u_h)_\Omega - \left(v_h, \delta \left[\beta \cdot \nabla\right] \left[\beta \cdot \nabla \right]u_h\right)_\Omega + \text{boundary terms}.
That means that the strong form of the equation would be of the sort
\beta \cdot \nabla u_h - \delta \left[\beta \cdot \nabla\right] \left[\beta \cdot \nabla \right] u_h.
What is important to recognize now is that \beta\cdot\nabla is the derivative in direction \beta. So, if we denote this by \beta\cdot\nabla=\frac{\partial}{\partial \beta} (in the same way as we often write \mathbf n\cdot\nabla=\frac{\partial}{\partial n} for the derivative in normal direction at the boundary), then the strong form of the equation is
\beta \cdot \nabla u_h - \delta \frac{\partial^2}{\partial\beta^2} u_h.
In other words, the unusual choice of test function is equivalent to the addition of term to the strong form that corresponds to a second order (i.e., diffusion) differential operator in the direction of the wind field \beta, i.e., in "streamline direction". A fuller account would also have to explore the effect of the test function on boundary values and why it is necessary to also use the same test function for the right hand side, but the discussion above might make clear where the name "streamline diffusion" for the method originates from.
A "Galerkin method" is one where one obtains the weak formulation by multiplying the equation by a test function v (and then integrating over \Omega) where the functions v are from the same space as the solution u (though possibly with different boundary values). But this is not strictly necessary: One could also imagine choosing the test functions from a different set of functions, as long as that different set has "as many dimensions" as the original set of functions so that we end up with as many independent equations as there are degrees of freedom (where all of this needs to be appropriately defined in the infinite-dimensional case). Methods that make use of this possibility (i.e., choose the set of test functions differently than the set of solutions) are called "Petrov-Galerkin" methods. In the current case, the test functions all have the form v+\beta\cdot\nabla v where v is from the set of solutions.
Upwind methods have a long history in the derivation of stabilized schemes for advection equations. Generally, the idea is that instead of looking at a function "here", we look at it a small distance further "upstream" or "upwind", i.e., where the information "here" originally came from. This might suggest not considering u(\mathbf x), but something like u(\mathbf x - \delta \beta). Or, equivalently upon integration, we could evaluate u(\mathbf x) and instead consider v a bit downstream: v(\mathbf x+\delta \beta). This would be cumbersome for a variety of reasons: First, we would have to define what v should be if \mathbf x + \delta \beta happens to be outside \Omega; second, computing integrals numerically would be much more awkward since we no longer evaluate u and v at the same quadrature points. But since we assume that \delta is small, we can do a Taylor expansion:
v(\mathbf x + \delta \beta) \approx v(\mathbf x) + \delta \beta \cdot \nabla v(\mathbf x).
This form for the test function should by now look familiar.
As the resulting matrix is no longer symmetric positive definite, we cannot use the usual Conjugate Gradient method (implemented in the SolverCG class) to solve the system. Instead, we use the GMRES (Generalized Minimum RESidual) method (implemented in SolverGMRES) that is suitable for problems of the kind we have here.
For the problem which we will solve in this tutorial program, we use the following domain and functions (in d=2 space dimensions):
\begin{eqnarray*} \Omega &=& [-1,1]^d \\ \beta({\mathbf x}) &=& \left( \begin{array}{c}2 \\ 1+\frac 45 \sin(8\pi x)\end{array} \right), \\ s &=& 0.1, \\ f({\mathbf x}) &=& \left\{ \begin{array}{ll} \frac 1{10 s^d} & \mathrm{for}\ |{\mathbf x}-{\mathbf x}_0|<s, \\ 0 & \mathrm{else}, \end{array} \right. \qquad\qquad {\mathbf x}_0 = \left( \begin{array}{c} -\frac 34 \\ -\frac 34\end{array} \right), \\ g &=& e^{5 (1 - |\mathbf x|^2)} \sin(16\pi|\mathbf x|^2). \end{eqnarray*}
For d>2, we extend \beta and {\mathbf x}_0 by simply duplicating the last of the components shown above one more time.
With all of this, the following comments are in order:
In all previous examples with adaptive refinement, we have used an error estimator first developed by Kelly et al., which assigns to each cell K the following indicator:
\eta_K = \left( \frac {h_K}{24} \int_{\partial K} [\partial_n u_h]^2 \; d\sigma \right)^{1/2},
where [\partial n u_h] denotes the jump of the normal derivatives across a face \gamma\subset\partial K of the cell K. It can be shown that this error indicator uses a discrete analogue of the second derivatives, weighted by a power of the cell size that is adjusted to the linear elements assumed to be in use here:
\eta_K \approx C h \| \nabla^2 u \|_K,
which itself is related to the error size in the energy norm.
The problem with this error indicator in the present case is that it assumes that the exact solution possesses second derivatives. This is already questionable for solutions to Laplace's problem in some cases, although there most problems allow solutions in H^2. If solutions are only in H^1, then the second derivatives would be singular in some parts (of lower dimension) of the domain and the error indicators would not reduce there under mesh refinement. Thus, the algorithm would continuously refine the cells around these parts, i.e. would refine into points or lines (in 2d).
However, for the present case, solutions are usually not even in H^1 (and this missing regularity is not the exceptional case as for Laplace's equation), so the error indicator described above is not really applicable. We will thus develop an indicator that is based on a discrete approximation of the gradient. Although the gradient often does not exist, this is the only criterion available to us, at least as long as we use continuous elements as in the present example. To start with, we note that given two cells K, K' of which the centers are connected by the vector {\mathbf y}_{KK'}, we can approximate the directional derivative of a function u as follows:
\frac{{\mathbf y}_{KK'}^T}{|{\mathbf y}_{KK'}|} \nabla u \approx \frac{u(K') - u(K)}{|{\mathbf y}_{KK'}|},
where u(K) and u(K') denote u evaluated at the centers of the respective cells. We now multiply the above approximation by {\mathbf y}_{KK'}/|{\mathbf y}_{KK'}| and sum over all neighbors K' of K:
\underbrace{ \left(\sum_{K'} \frac{{\mathbf y}_{KK'} {\mathbf y}_{KK'}^T} {|{\mathbf y}_{KK'}|^2}\right)}_{=:Y} \nabla u \approx \sum_{K'} \frac{{\mathbf y}_{KK'}}{|{\mathbf y}_{KK'}|} \frac{u(K') - u(K)}{|{\mathbf y}_{KK'}|}.
If the vectors {\mathbf y}_{KK'} connecting K with its neighbors span the whole space (i.e. roughly: K has neighbors in all directions), then the term in parentheses in the left hand side expression forms a regular matrix, which we can invert to obtain an approximation of the gradient of u on K:
\nabla u \approx Y^{-1} \left( \sum_{K'} \frac{{\mathbf y}_{KK'}}{|{\mathbf y}_{KK'}|} \frac{u(K') - u(K)}{|{\mathbf y}_{KK'}|} \right).
We will denote the approximation on the right hand side by \nabla_h u(K), and we will use the following quantity as refinement criterion:
\eta_K = h^{1+d/2} |\nabla_h u_h(K)|,
which is inspired by the following (not rigorous) argument:
\begin{eqnarray*} \|u-u_h\|^2_{L_2} &\le& C h^2 \|\nabla u\|^2_{L_2} \\ &\approx& C \sum_K h_K^2 \|\nabla u\|^2_{L_2(K)} \\ &\le& C \sum_K h_K^2 h_K^d \|\nabla u\|^2_{L_\infty(K)} \\ &\approx& C \sum_K h_K^{2+d} |\nabla_h u_h(K)|^2 \end{eqnarray*}
Just as in previous examples, we have to include several files of which the meaning has already been discussed:
The following two files provide classes and information for multithreaded programs. In the first one, the classes and functions are declared which we need to do assembly in parallel (i.e. the WorkStream namespace). The second file has a class MultithreadInfo which can be used to query the number of processors in your system, which is often useful when deciding how many threads to start in parallel.
The next new include file declares a base class TensorFunction not unlike the Function class, but with the difference that TensorFunction::value returns a Tensor instead of a scalar.
This is C++, as we want to write some output to disk:
The last step is as in previous programs:
Next we declare a class that describes the advection field. This, of course, is a vector field with as many components as there are space dimensions. One could now use a class derived from the Function base class, as we have done for boundary values and coefficients in previous examples, but there is another possibility in the library, namely a base class that describes tensor valued functions. This is more convenient than overriding Function::value() with a method that knows about multiple function components: At the end of the day we need a Tensor, so we may as well just use a class that returns a Tensor.
Besides the advection field, we need two functions describing the source terms (right hand side) and the boundary values. As described in the introduction, the source is a constant function in the vicinity of a source point, which we denote by the constant static variable center_point. We set the values of this center using the same template tricks as we have shown in the step-7 example program. The rest is simple and has been shown previously.
The only new thing here is that we check for the value of the component parameter. As this is a scalar function, it is obvious that it only makes sense if the desired component has the index zero, so we assert that this is indeed the case. ExcIndexRange is a global predefined exception (probably the one most often used, we therefore made it global instead of local to some class), that takes three parameters: the index that is outside the allowed range, the first element of the valid range and the one past the last (i.e. again the half-open interval so often used in the C++ standard library):
Finally for the boundary values, which is just another class derived from the Function base class:
Here comes the main class of this program. It is very much like the main classes of previous examples, so we again only comment on the differences.
The next set of functions will be used to assemble the matrix. However, unlike in the previous examples, the assemble_system() function will not do the work itself, but rather will delegate the actual assembly to helper functions assemble_local_system() and copy_local_to_global(). The rationale is that matrix assembly can be parallelized quite well, as the computation of the local contributions on each cell is entirely independent of other cells, and we only have to synchronize when we add the contribution of a cell to the global matrix.
The strategy for parallelization we choose here is one of the possibilities mentioned in detail in the Parallel computing with multiple processors accessing shared memory topic in the documentation. Specifically, we will use the WorkStream approach discussed there. Since there is so much documentation in this group, we will not repeat the rationale for the design choices here (for example, if you read through the topic mentioned above, you will understand what the purpose of the AssemblyScratchData and AssemblyCopyData structures is). Rather, we will only discuss the specific implementation.
If you read the page mentioned above, you will find that in order to parallelize assembly, we need two data structures – one that corresponds to data that we need during local integration ("scratch data", i.e., things we only need as temporary storage), and one that carries information from the local integration to the function that then adds the local contributions to the corresponding elements of the global matrix. The former of these typically contains the FEValues and FEFaceValues objects, whereas the latter has the local matrix, local right hand side, and information about which degrees of freedom live on the cell for which we are assembling a local contribution. With this information, the following should be relatively self-explanatory:
FEValues and FEFaceValues are expensive objects to set up, so we include them in the scratch object so that as much data is reused between cells as possible.
We also store a few vectors that we will populate with values on each cell. Setting these objects up is, in the usual case, cheap; however, they require memory allocations, which can be expensive in multithreaded applications. Hence we keep them here so that computations on a cell do not require new allocations.
Finally, we need objects that describe the problem's data:
The following functions again are the same as they were in previous examples, as are the subsequent variables:
Now, finally, here comes the class that will compute the difference approximation of the gradient on each cell and weighs that with a power of the mesh size, as described in the introduction. This class is a simple version of the DerivativeApproximation class in the library, that uses similar techniques to obtain finite difference approximations of the gradient of a finite element field, or of higher derivatives.
The class has one public static function estimate that is called to compute a vector of error indicators, and a few private functions that do the actual work on all active cells. As in other parts of the library, we follow an informal convention to use vectors of floats for error indicators rather than the common vectors of doubles, as the additional accuracy is not necessary for estimated values.
In addition to these two functions, the class declares two exceptions which are raised when a cell has no neighbors in each of the space directions (in which case the matrix described in the introduction would be singular and can't be inverted), while the other one is used in the more common case of invalid parameters to a function, namely a vector of wrong size.
Two other comments: first, the class has no non-static member functions or variables, so this is not really a class, but rather serves the purpose of a namespace in C++. The reason that we chose a class over a namespace is that this way we can declare functions that are private. This can be done with namespaces as well, if one declares some functions in header files in the namespace and implements these and other functions in the implementation file. The functions not declared in the header file are still in the namespace but are not callable from outside. However, as we have only one file here, it is not possible to hide functions in the present case.
The second comment is that the dimension template parameter is attached to the function rather than to the class itself. This way, you don't have to specify the template parameter yourself as in most other cases, but the compiler can figure its value out itself from the dimension of the DoFHandler object that one passes as first argument.
Before jumping into the fray with the implementation, let us also comment on the parallelization strategy. We have already introduced the necessary framework for using the WorkStream concept in the declaration of the main class of this program above. We will use it again here. In the current context, this means that we have to define
Given this general framework, we will, however, deviate from it a bit. In particular, WorkStream was generally invented for cases where each local computation on a cell adds to a global object – for example, when assembling linear systems where we add local contributions into a global matrix and right hand side. WorkStream is designed to handle the potential conflict of multiple threads trying to do this addition at the same time, and consequently has to provide for some way to ensure that only one thread gets to do this at a time. Here, however, the situation is slightly different: we compute contributions from every cell individually, but then all we need to do is put them into an element of an output vector that is unique to each cell. Consequently, there is no risk that the write operations from two cells might conflict, and the elaborate machinery of WorkStream to avoid conflicting writes is not necessary. Consequently, what we will do is this: We still need a scratch object that holds, for example, the FEValues object. However, we only create a fake, empty copy data structure. Likewise, we do need the function that computes local contributions, but since it can already put the result into its final location, we do not need a copy-local-to-global function and will instead give the WorkStream::run() function an empty function object – the equivalent to a NULL function pointer.
There is one more part to this class. In previous examples, we have used assertions that throw exceptions in several places (see, for example, step-5 and step-8). However, you have never seen how such exceptions are declared. The code below shows how to do that, using the DeclException2 macro.
The syntax may look a little strange, but is reasonable. The format is basically as follows: Use the name of one of the macros DeclExceptionN, where N denotes the number of additional parameters which the exception object should take. In this case, as we want to throw the exception when the sizes of two vectors differ, we need two arguments, so we use DeclException2. The first parameter then describes the name of the exception, while the following declare the data types of the parameters. The last argument is a sequence of output directives that will be piped into the std::cerr object, thus the strange format with the leading << operator and the like. Note that we can access the parameters which are passed to the exception upon construction (i.e. within the Assert call) by using the names arg1 through argN, where N is the number of arguments as defined by the use of the respective macro DeclExceptionN.
To learn how the preprocessor expands this macro into actual code, please refer to the documentation of the exception classes. In brief, this macro call declares and defines a class ExcDimensionMismatch inheriting from ExceptionBase which implements all necessary error output functions.
AssertDimension macro, which is a handy wrapper for checking that the dimensions of two given objects are equal, and that you have already seen in step-8.Now for the implementation of the main class. Constructor, destructor and the function setup_system follow the same pattern that was used previously, so we need not comment on these three function:
In the following function, the matrix and right hand side are assembled. As stated in the documentation of the main class above, it does not do this itself, but rather delegates to the function following next, utilizing the WorkStream concept discussed in Parallel computing with multiple processors accessing shared memory .
If you have looked through the Parallel computing with multiple processors accessing shared memory topic, you will have seen that assembling in parallel does not take an incredible amount of extra code as long as you diligently describe what the scratch and copy data objects are, and if you define suitable functions for the local assembly and the copy operation from local contributions to global objects. This done, the following will do all the heavy lifting to get these operations done on multiple threads on as many cores as you have in your system:
As already mentioned above, we need to have scratch objects for the parallel computation of local contributions. These objects contain FEValues and FEFaceValues objects (as well as some arrays), and so we will need to have constructors and copy constructors that allow us to create them. For the cell terms we need the values and gradients of the shape functions, the quadrature points in order to determine the source density and the advection field at a given point, and the weights of the quadrature points times the determinant of the Jacobian at these points. In contrast, for the boundary integrals, we don't need the gradients, but rather the normal vectors to the cells. This determines which update flags we will have to pass to the constructors of the members of the class:
Now, this is the function that does the actual work. It is not very different from the assemble_system functions of previous example programs, so we will again only comment on the differences. The mathematical stuff closely follows what we have said in the introduction.
There are a number of points worth mentioning here, though. The first one is that we have moved the FEValues and FEFaceValues objects into the ScratchData object. We have done so because the alternative would have been to simply create one every time we get into this function – i.e., on every cell. It now turns out that the FEValues classes were written with the explicit goal of moving everything that remains the same from cell to cell into the construction of the object, and only do as little work as possible in FEValues::reinit() whenever we move to a new cell. What this means is that it would be very expensive to create a new object of this kind in this function as we would have to do it for every cell – exactly the thing we wanted to avoid with the FEValues class. Instead, what we do is create it only once (or a small number of times) in the scratch objects and then re-use it as often as we can.
This begs the question of whether there are other objects we create in this function whose creation is expensive compared to its use. Indeed, at the top of the function, we declare all sorts of objects. The AdvectionField, RightHandSide and BoundaryValues do not cost much to create, so there is no harm here. However, allocating memory in creating the rhs_values and similar variables below typically costs a significant amount of time, compared to just accessing the (temporary) values we store in them. Consequently, these would be candidates for moving into the AssemblyScratchData class. We will leave this as an exercise.
We define some abbreviations to avoid unnecessarily long lines:
We declare cell matrix and cell right hand side...
... an array to hold the global indices of the degrees of freedom of the cell on which we are presently working...
... then initialize the FEValues object...
... obtain the values of right hand side and advection directions at the quadrature points...
... set the value of the streamline diffusion parameter as described in the introduction...
... and assemble the local contributions to the system matrix and right hand side as also discussed above:
Alias the AssemblyScratchData object to keep the lines from getting too long:
Besides the cell terms which we have built up now, the bilinear form of the present problem also contains terms on the boundary of the domain. Therefore, we have to check whether any of the faces of this cell are on the boundary of the domain, and if so assemble the contributions of this face as well. Of course, the bilinear form only contains contributions from the inflow part of the boundary, but to find out whether a certain part of a face of the present cell is part of the inflow boundary, we have to have information on the exact location of the quadrature points and on the direction of flow at this point; we obtain this information using the FEFaceValues object and only decide within the main loop whether a quadrature point is on the inflow boundary.
Ok, this face of the present cell is on the boundary of the domain. Just as for the usual FEValues object which we have used in previous examples and also above, we have to reinitialize the FEFaceValues object for the present face:
For the quadrature points at hand, we ask for the values of the inflow function and for the direction of flow:
Now loop over all quadrature points and see whether this face is on the inflow or outflow part of the boundary. The normal vector points out of the cell: since the face is at the boundary, the normal vector points out of the domain, so if the advection direction points into the domain, its scalar product with the normal vector must be negative (to see why this is true, consider the scalar product definition that uses a cosine):
If the face is part of the inflow boundary, then compute the contributions of this face to the global matrix and right hand side, using the values obtained from the FEFaceValues object and the formulae discussed in the introduction:
The final piece of information the copy routine needs is the global indices of the degrees of freedom on this cell, so we end by writing them to the local array:
The second function we needed to write was the one that copies the local contributions the previous function computed (and put into the AssemblyCopyData object) into the global matrix and right hand side vector objects. This is essentially what we always had as the last block of code when assembling something on every cell. The following should therefore be pretty obvious:
The next function is the linear solver routine. As the system is no longer symmetric positive definite as in all the previous examples, we cannot use the Conjugate Gradient method any more. Rather, we use a solver that is more general and does not rely on any special properties of the matrix: the GMRES method. GMRES, like the conjugate gradient method, requires a decent preconditioner: we use a Jacobi preconditioner here, which works well enough for this problem.
The following function refines the grid according to the quantity described in the introduction. The respective computations are made in the class GradientEstimation.
This function is similar to the one in step-6, but since we use a higher degree finite element we save the solution in a different way. Visualization programs like VisIt and Paraview typically only understand data that is associated with nodes: they cannot plot fifth-degree basis functions, which results in a very inaccurate picture of the solution we computed. To get around this we save multiple patches per cell: in 2d we save 8\times 8=64 bilinear ‘sub-cells’ to the VTU file for each cell, and in 3d we save 8\times 8\times 8 = 512. The end result is that the visualization program will use a piecewise linear interpolation of the cubic basis functions on a 3 times refined mesh: This captures the solution detail and, with most screen resolutions, looks smooth. We save the grid in a separate step with no extra patches so that we have a visual representation of the cell faces.
VTU output can be expensive, both to compute and to write to disk. Here we ask ZLib, a compression library, to compress the data in a way that maximizes throughput.
... as is the main loop (setup – solve – refine), aside from the number of cycles and the initial grid:
Now for the implementation of the GradientEstimation class. Let us start by defining constructors for the EstimateScratchData class used by the estimate_cell() function:
We allocate a vector to hold iterators to all active neighbors of a cell. We reserve the maximal number of active neighbors in order to avoid later reallocations. Note how this maximal number of active neighbors is computed here.
Next comes the implementation of the GradientEstimation class. The first function does not much except for delegating work to the other function, but there is a bit of setup at the top.
Before starting with the work, we check that the vector into which the results are written has the right size, using the Assert macro and the exception class we declared above. Programming mistakes in which one forgets to size arguments correctly at the calling site are quite common. Because the resulting damage from not catching such errors is often subtle (e.g., corruption of data somewhere in memory, or non-reproducible results), it is well worth the effort to check for such things.
Here comes the function that estimates the local error by computing the finite difference approximation of the gradient. The function first computes the list of active neighbors of the present cell and then computes the quantities described in the introduction for each of the neighbors. The reason for this order is that it is not a one-liner to find a given neighbor with locally refined meshes. In principle, an optimized implementation would find neighbors and the quantities depending on them in one step, rather than first building a list of neighbors and in a second step their contributions but we will gladly leave this as an exercise. As discussed before, the worker function passed to WorkStream::run works on "scratch" objects that keep all temporary objects. This way, we do not need to create and initialize objects that are expensive to initialize within the function that does the work every time it is called for a given cell. Such an argument is passed as the second argument. The third argument would be a "copy-data" object (see Parallel computing with multiple processors accessing shared memory for more information) but we do not actually use any of these here. Since WorkStream::run() insists on passing three arguments, we declare this function with three arguments, but simply ignore the last one.
(This is unsatisfactory from an aesthetic perspective. It can be avoided by using an anonymous (lambda) function. If you allow, let us here show how. First, assume that we had declared this function to only take two arguments by omitting the unused last one. Now, WorkStream::run still wants to call this function with three arguments, so we need to find a way to "forget" the third argument in the call. Simply passing WorkStream::run the pointer to the function as we do above will not do this – the compiler will complain that a function declared to have two arguments is called with three arguments. However, we can do this by passing the following as the third argument to WorkStream::run():
This is not much better than the solution implemented below: either the routine itself must take three arguments or it must be wrapped by something that takes three arguments. We don't use this since adding the unused argument at the beginning is simpler.
Now for the details:
We need space for the tensor Y, which is the sum of outer products of the y-vectors.
First initialize the FEValues object, as well as the Y tensor:
Now, before we go on, we first compute a list of all active neighbors of the present cell. We do so by first looping over all faces and see whether the neighbor there is active, which would be the case if it is on the same level as the present cell or one level coarser (note that a neighbor can only be once coarser than the present cell, as we only allow a maximal difference of one refinement over a face in deal.II). Alternatively, the neighbor could be on the same level and be further refined; then we have to find which of its children are next to the present cell and select these (note that if a child of a neighbor of an active cell that is next to this active cell, needs necessarily be active itself, due to the one-refinement rule cited above).
Things are slightly different in one space dimension, as there the one-refinement rule does not exist: neighboring active cells may differ in as many refinement levels as they like. In this case, the computation becomes a little more difficult, but we will explain this below.
Before starting the loop over all neighbors of the present cell, we have to clear the array storing the iterators to the active neighbors, of course.
First define an abbreviation for the iterator to the face and the neighbor
Then check whether the neighbor is active. If it is, then it is on the same level or one level coarser (if we are not in 1d), and we are interested in it in any case.
If the neighbor is not active, then check its children.
To find the child of the neighbor which bounds to the present cell, successively go to its right child if we are left of the present cell (n==0), or go to the left child if we are on the right (n==1), until we find an active cell.
As this used some non-trivial geometrical intuition, we might want to check whether we did it right, i.e., check whether the neighbor of the cell we found is indeed the cell we are presently working on. Checks like this are often useful and have frequently uncovered errors both in algorithms like the line above (where it is simple to involuntarily exchange n==1 for n==0 or the like) and in the library (the assumptions underlying the algorithm above could either be wrong, wrongly documented, or are violated due to an error in the library). One could in principle remove such checks after the program works for some time, but it might be a good things to leave it in anyway to check for changes in the library or in the algorithm above.
Note that if this check fails, then this is certainly an error that is irrecoverable and probably qualifies as an internal error. We therefore use a predefined exception class to throw here.
If the check succeeded, we push the active neighbor we just found to the stack we keep:
If we are not in 1d, we collect all neighbor children ‘behind’ the subfaces of the current face and move on:
OK, now that we have all the neighbors, lets start the computation on each of them. First we do some preliminaries: find out about the center of the present cell and the solution at this point. The latter is obtained as a vector of function values at the quadrature points, of which there are only one, of course. Likewise, the position of the center is the position of the first (and only) quadrature point in real space.
Now loop over all active neighbors and collect the data we need.
Then get the center of the neighbor cell and the value of the finite element function at that point. Note that for this information we have to reinitialize the FEValues object for the neighbor cell.
Compute the vector y connecting the centers of the two cells. Note that as opposed to the introduction, we denote by y the normalized difference vector, as this is the quantity used everywhere in the computations.
Then add up the contribution of this cell to the Y matrix...
... and update the sum of difference quotients:
If now, after collecting all the information from the neighbors, we can determine an approximation of the gradient for the present cell, then we need to have passed over vectors y which span the whole space, otherwise we would not have all components of the gradient. This is indicated by the invertibility of the matrix.
If the matrix is not invertible, then the present cell had an insufficient number of active neighbors. In contrast to all previous cases (where we raised exceptions) this is, however, not a programming error: it is a runtime error that can happen in optimized mode even if it ran well in debug mode, so it is reasonable to try to catch this error also in optimized mode. For this case, there is the AssertThrow macro: it checks the condition like the Assert macro, but not only in debug mode; it then outputs an error message, but instead of aborting the program as in the case of the Assert macro, the exception is thrown using the throw command of C++. This way, one has the possibility to catch this error and take reasonable counter actions. One such measure would be to refine the grid globally, as the case of insufficient directions can not occur if every cell of the initial grid has been refined at least once.
If, on the other hand, the matrix is invertible, then invert it, multiply the other quantity with it, and compute the estimated error using this quantity and the correct powers of the mesh width:
The last part of this function is the one where we write into the element of the output vector what we have just computed. The address of this vector has been stored in the scratch data object, and all we have to do is know how to get at the correct element inside this vector – but we can ask the cell we're on the how-manyth active cell it is for this:
The main function is similar to the previous examples. The primary difference is that we use MultithreadInfo to set the maximum number of threads (see the documentation topic Parallel computing with multiple processors accessing shared memory for more information). The number of threads used is the minimum of the environment variable DEAL_II_NUM_THREADS and the parameter of set_thread_limit. If no value is given to set_thread_limit, the default value from the Intel Threading Building Blocks (TBB) library is used. If the call to set_thread_limit is omitted, the number of threads will be chosen by TBB independently of DEAL_II_NUM_THREADS.
The results of this program are not particularly spectacular. They consist of the console output, some grid files, and the solution on each of these grids. First for the console output:
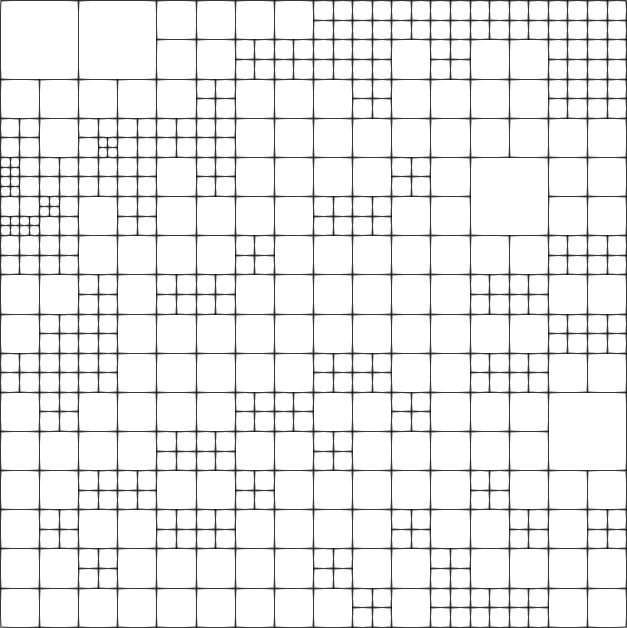
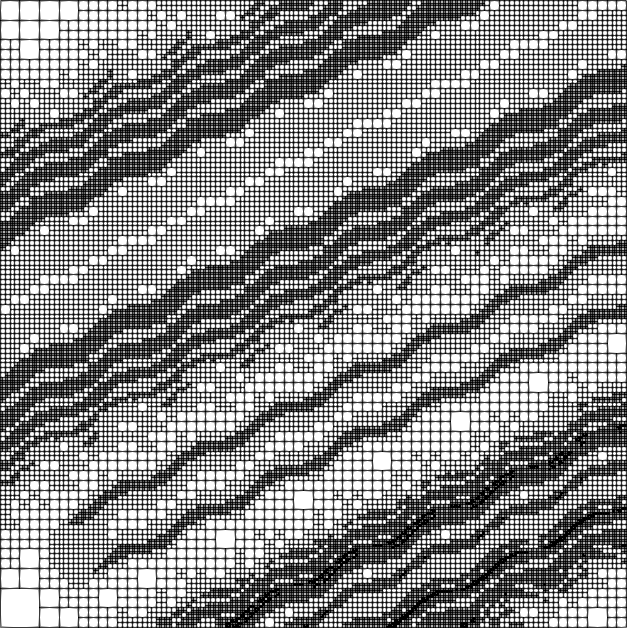
Quite a number of cells are used on the finest level to resolve the features of the solution. Here are the fourth and tenth grids:
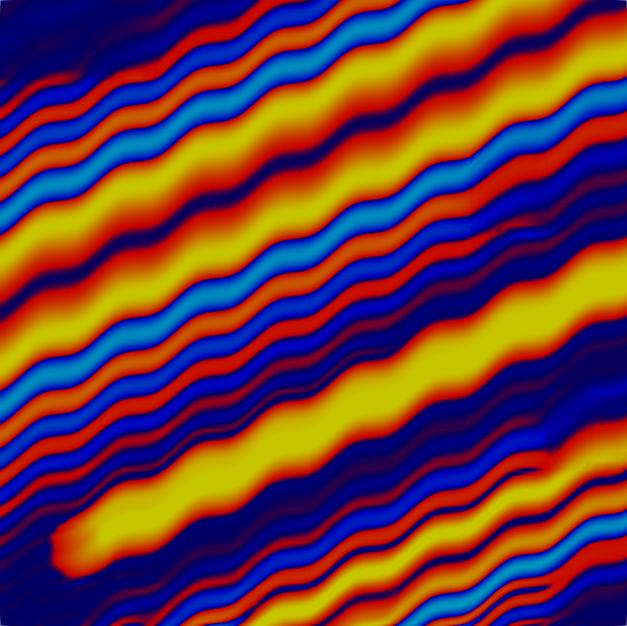
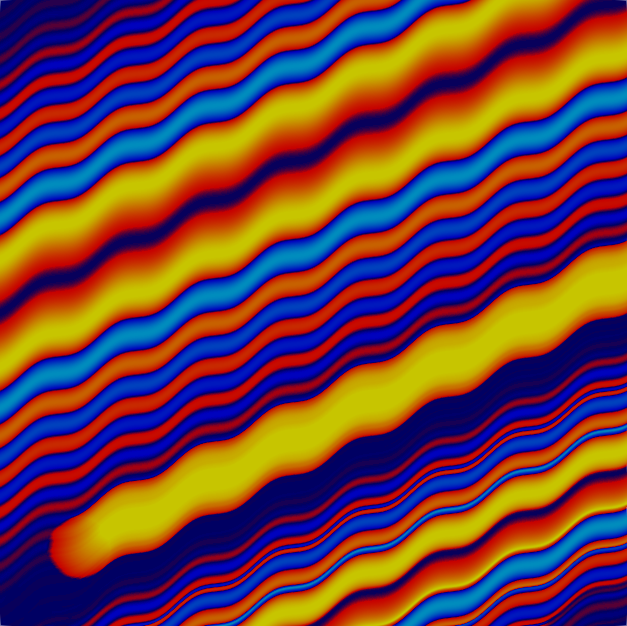
and the fourth and tenth solutions:
and both the grid and solution zoomed in:
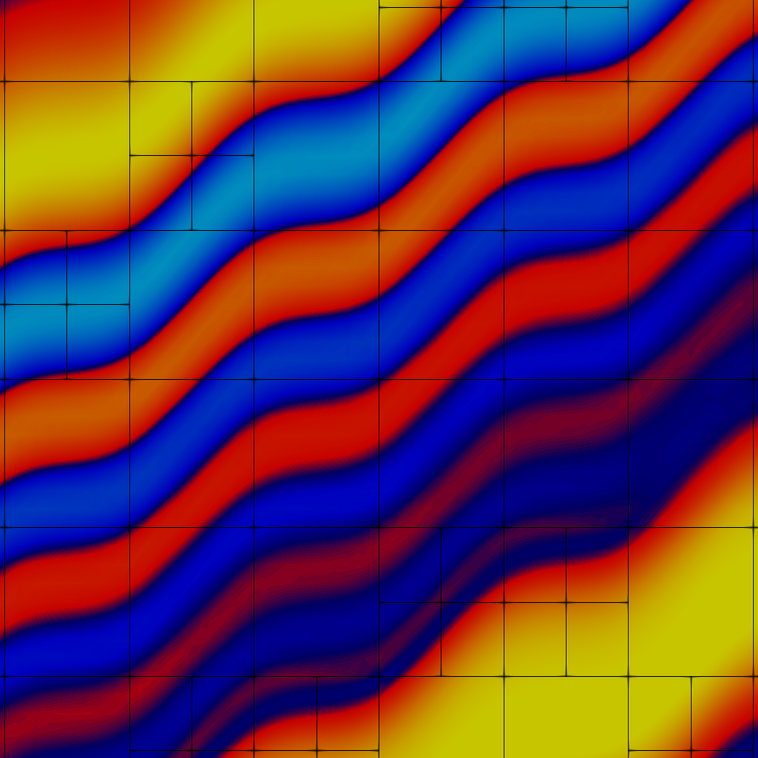
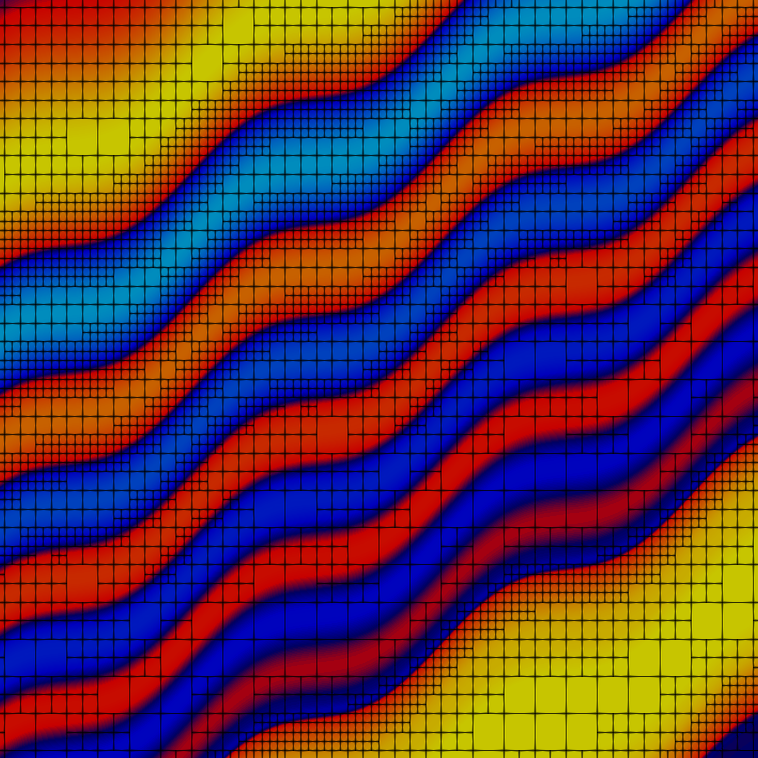
The solution is created by that part that is transported along the wiggly advection field from the left and lower boundaries to the top right, and the part that is created by the source in the lower left corner, and the results of which are also transported along. The grid shown above is well-adapted to resolve these features. The comparison between plots shows that, even though we are using a high-order approximation, we still need adaptive mesh refinement to fully resolve the wiggles.