This program was contributed by Wolfgang Bangerth <bangerth@colostate.edu>.
It comes without any warranty or support by its authors or the authors of deal.II.
This program is part of the deal.II code gallery and consists of the following files (click to inspect):
Pictures from this code gallery program
Annotated version of Matlab/README.md
Bayesian inversion benchmark: Matlab version
This code implements a Matlab version of the benchmark in D. Aristoff and W. Bangerth, "A benchmark for the Bayesian inversion of coefficients in partial differential equations," arXiv:2102.07263. The article is available on the web at https://arxiv.org/abs/2102.07263.
The benchmark is intended for comparing advanced methods for Bayesian inverse problems. The benchmark is designed to be more complex than simple testcases like Gaussian mixtures, but not so complex as to be intractable or unrepeatable. The benchmark concerns the determination of a spatially variable coefficient, discretized by 64 values, in a Poisson equation, based on point measurements of the solution.
This code runs a basic Metropolis-Hastings sampler for the posterior distribution. To run it, put all the .m and the .mat files from this repository in your current Matlab directory, and type "main" into the Matlab command window. This starts the main program, main.m, which lets you can choose a number of independent Markov chains, the length of each chain, the lag time between recording samples, and the number of parallel workers to use. Other parameters for the simulations can be found within precomputations.m.
The posterior distribution has argument theta, which is a vector of length 64 representing the spatially variable coefficient in the Poisson equation. This vector represents the values of the coefficient in a 8x8 grid. In this code, theta is represented by an 8x8 matrix. To convert a vector, theta, of length 64 – with entries associated to coefficients as described in the article above – into an equivalent 8x8 matrix suitable for this code, use the Matlab command "theta = reshape(theta,[8 8])';". The matrix of exact coefficients can be obtained using the command "theta = ones(8,8); theta(2:3,2:3) = 0.1; theta(6:7,6:7) = 10;".
Annotated version of Readme.md
Readme file for MCMC-Laplace
- Note
- The intent and implementation of this program is extensively described in D. Aristoff and W. Bangerth: "A benchmark for the Bayesian
inversion of coefficients in partial differential equations", submitted, 2021. A preprint can be found here. See there for more information.
Motivation for project
Inverse problems are problems in which one (typically) wants to infer something about the internal properties of a body by measuring how it reacts to an external stimulus. An example would be that you want to determine the stiffness parameters of a membrane by applying an external force to it and measuring how it deforms. A more complicated inverse problem is determining the three-dimensional make-up of the Earth by measuring the time it takes for seismic waves to travel from the source of an Earthquake to far-away detectors. Most biomedical imaging techniques are also inverse problems.
The traditional approach to inverse problems is to ask the question which hypothesized make-up of the body would result in predicted reactions that are "closest" to the measured one. This formulation of the problem is what is now generally called the "deterministic inverse
problem", and it is an optimization problem: Among all possible make-ups of the body, find the one which minimizes the difference between predicted measurements and actual measurements.
Since the late 1990s, a second paradigm for the formulation has come into play: "Bayesian inverse problems". It rests on the observation that our measurements are not exact but rather that certain values are just more or less likely to show up on the dial of the instrument we measure with. For example, if a device measures the deformation of a membrane as 2.85 cm, and if we know that the measuring device has a Gaussian-distributed uncertainty with standard deviation 0.05 cm, then the Bayesian inverse problem asks for finding a probability distribution among all of the make-ups of the body so that the predicted measurements have the observed distribution of a Gaussian with mean 2.85 cm and standard deviation 0.05 cm.
To make things more concrete, let us denote the parameters that describe the internal make-up of the membrane as the vector \(\mathbf
a\), and the measured deflections at a set of measurement points as \(\mathbf z\). Assume that we have measured a set of values \(\hat
{\mathbf z}\), and that we know that each of these measurements is normal distributed with standard deviation \(\sigma\), i.e., that the "real" values are \(\mathbf z \sim N(\hat {\mathbf z}, \sigma I)\) – i.e., normally distributed with mean \(\hat {\mathbf z}\) and covariance matrix \(\sigma
I\).
Let us further assume that for each set of parameters \(\mathbf a\), we can predict measurements \(\mathbf z=\mathbf F(\mathbf a)\) with some function \(\mathbf F(\cdot)\) that in general will involve solving a partial differential equation with known external applied force and given trial coefficients \(\mathbf a\). What we are interested in is what the probability distribution \(\pi(\mathbf a)\) is so that the corresponding \(\pi(\mathbf z)=\pi(\mathbf F(\mathbf a))=N(\hat{\mathbf
z},\sigma I)\). This problem can, in general, not be solved exactly because we only know \(\mathbf F\), the parameters-to-measurements map that can be evaluated by solving the PDE and then evaluating the solution at individual points, but not the inverse of \(\mathbf
F\). But, it is possible to sample from the distribution \(\pi(\mathbf
a)\) using Monte Carlo Markov Chain (MCMC) methods.
This is what this program does, in essence. The formulation of the problem is marginally more complicated than outlined above, also taking into account a prior distribution that describes some assumptions we may have on the parameter. But in essence, this is what we are doing:
- There is a Metropolis-Hastings that implements a Markov Chain to sample from \(\pi(\mathbf a)\). The Markov chain that results from this is then a (very long) sequence of samples \(\mathbf a_1, \mathbf
a_2, \ldots\) that are written to a (potentially very large) output file. If you don't know what a Metropolis-Hastings sampler is, or how a sequence of samples approximates a probability distribution, then you will probably want to take a look at the Wikipedia pages for Markov Chain Monte Carlo methods and for the Metropolis-Hastings algorithm.
- As part of the sampling process, we need to solve the PDE that describes the physical system here.
- The remainder of the program is devoted to the description of the distribution \(\pi(\mathbf z)\) we provide as input, as well as a number of other pieces of information that enter into the definition of what exactly the Metropolis-Hastings sampler does here.
- Note
- This program computes samples the brute force way. We want to compute billions of samples using a simple algorithm because we want a benchmark that smarter algorithms can be tested again. The point isn't that the Metropolis-Hastings algorithm as implemented here (using, in particular, an isotropic proposal distribution) is the sharpest tool in the shed – it isn't – but that it is reliable and robust. We want to see what it converges to, and how fast, so that we can test better sampling methods against this baseline.
More detailed properties of the code in MCMC-Laplace
To be concise, the problem we are considering is the following: We are assuming that the membrane we are deforming through an external force is a square with edge length 1 (i.e., the domain is \(\Omega=(0,1)^2\)) and that it is made up of \(8\times 8\) smaller squares each of which has a constant stiffness \(a_k,
k=0,\ldots,63\). In other words, we would like to find the vector \(\mathbf a=(a_0,\ldots,a_{63})^T\) for which the predicted deformation matches our measurements \(\hat{\mathbf z}\) in the sense discussed above.
The model of deformation we consider is the Poisson equation with a non-constant coefficient:
\begin{align*}
-\nabla \cdot (a(\mathbf x) \nabla u(\mathbf x) &= f(\mathbf x)
\qquad\qquad &&\text{in}\ \Omega,
\\
u(\mathbf x) &= 0
\qquad\qquad &&\text{on}\ \partial\Omega.
\end{align*}
Here, the spatially variable coefficient \(a(\mathbf x)\) corresponds to the 64 values in \(\mathbf a\) by mapping the elements of \(\mathbf a\) to regions of the mesh. We choose \(f=10\), which results in a solution that is approximately equal to one at its maximum. The following picture shows this solution \(u\): 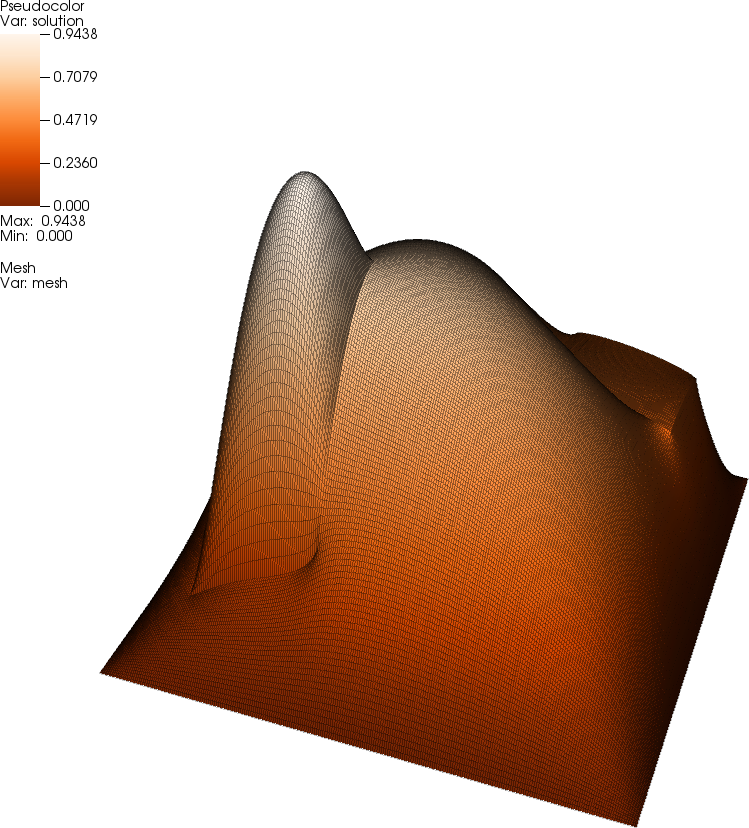 The coefficient values that correspond to this solution (the "exact" coefficient from which the measurements \(\hat{\mathbf z}\) were generated) looks as follows:
The coefficient values that correspond to this solution (the "exact" coefficient from which the measurements \(\hat{\mathbf z}\) were generated) looks as follows: 
For every given coefficient \(\mathbf a\), the corresponding measurement values \(z_i, i=0,\ldots,168\) are then obtained by evaluating the solution \(u\) on a \(13\times 13\) grid of equidistance points \(\mathbf
x_i\).
You will find these concepts mapped into the code as part of the PoissonSolver class. Of particular interest may be the fact that the computation of \(\mathbf z\) by evaluating \(u\) at individual points is a linear operation, and consequently can be represented using a matrix applied to the solution vector. (In the code, this corresponds to the PoissonSolver::measurement_matrix member variable.) Furthermore, we make the assumption that the mesh used in solving the PDE is at least as fine as the \(8\times 8\) mesh used to represent the coefficient \(\mathbf a\) we would like to infer; then, the coefficient is constant on each cell, and we can get the value of the coefficient on a given cell by looking up the corresponding value of the element of the vector \(\mathbf a\). We store the index of this vector element in the user_index property that deal.II provides for each cell, and set this connection up in PoissonSolver::setup_system().
The only other part worth discussing about this program is that it is set up for speed. This program implementing a benchmark, we are interested generating as many samples as possible – the paper mentioned at the top of this page shows data obtained from more than \(10^{10}\) samples. To compute this many samples, solving the PDE cannot take too long or we would never finish the paper. The question then is how, given a set of coefficients \(\mathbf a\), we can assemble and solve the linear systems for the Poisson equation as quickly as possible. In the current program, this is done using the observation that the local contribution to the global matrix is simply a matrix that is the same for every cell (because we are using a mesh in which every cell looks the same) times the coefficient for the current cell. This is because we know that the coefficient is constant on every cell, as discussed above. As a consequence, we compute the local matrix (with a unit coefficient) only once, in PoissonProblem::setup_system(), using the very first cell. We do the same with the local right hand side vector, which is again the same for every cell because the right hand side function is constant.
During assembly of the linear system, we then only need to recall these local matrix and right hand side contributions, multiply the local matrix by the coefficient of the current cell, and then copy everything into the global matrix as usual.
To run the code
After running cmake and compiling via make (or, if you have used the -G ... option of cmake, compiling the program via your favorite integrated development environment), you can run the executable by either just saying make run or using ./mcmc-laplace on the command line. The default is to compile in "debug mode"; you can switch to "release mode" by saying make release and then compiling everything again.
The program as is will run in around 40 seconds on a current machine at the time of writing this program when compiled in release mode. This is in the test mode that is the default setting selected in the main() function, and it produces 10,000 samples. This is enough to get an idea of what the program does. For real simulations, such as those discussed in the paper referenced at the top, one of course wants to have many many more samples; if you select testing = false at the top of main(), the program will create 100,000,000 samples, which will take around a day and a half to run in release mode. That may be more than you've bargained for, but you can always terminate the program, or just select a smaller number of samples at the bottom of main().
When not in testing mode, the program initializes all random number generators that are part of the Metropolis-Hastins algorithms with a seed that is created using the std::random_device() function, a function that uses the operating system to create a seed that may take into account the current time, the amount of data written to disk over the past hour, the amount of internet traffic that has gone through the machine in the last hour, and similar pieces of pretty much random information. As a consequence, the seed is then pretty much guaranteed to be different from program invokation to program invokation, and consequently we will get different random number sequences every time. The output file is tagged with a string representation of this random seed, so that it is safe to run the same program multiple times at the same time in the same directory, with each running program writing a different sequence of samples into separate files.
The end result of the program is a file that contains the samples. Each line has 66 entries:
- The first entry is the logarithm of the (non-normalized) posterior probability of the sample; because the posterior is only known up to a normalization constant, the absolute value is not relevant, but the relative values of different samples are informative.
- The second entry is the number of samples accepted up to this point. By counting how many lines one is into a given file (i.e., counting the total number of samples up to this point), this number is useful to compute the acceptance rate of the Metropolis-Hastings algorithm.
- The remaining 64 numbers are the entries of the current sample vector.
Testing modifications and alternative implementations
In order to verify that modifications made to this code are correct, or that alternative implementations of the benchmark in other programming languages produce the expected results, it is useful to have known outputs for certain inputs. To this end, the testing/ directory contains ten files with 64-dimensional input vectors: Each of the files testing/input.X.txt contains a vector of coefficients (theta) for which the corresponding testing/output.X.z.txt contains the corresponding output at the evaluation points (the z vector that corresponds to the theta vector, as described in the paper linked to at the very top of this page). Furthermore, the testing/output.X.likelihood.txt file contains the corresponding likelihood value for this theta vector that can be computed from the z vector, and the testing/output.X.prior.txt file contains the prior associated with this theta vector. The values in these last two files are not normalized, and so care must be taken when comparing these values between implementations: An implementation (or a patched version of this program) may compute a different value, but the ratio of the values between different inputs must be the same – in other words, the outputs must be like the ones stored in these files up to a constant for an implementation to be correct.
The ten inputs are chosen as follows:
The testing/input.0.txt file corresponds to a 64-dimensional vector of all ones. The testing/input.1.txt file corresponds to a 64-dimensional vector of coefficients theta that are all equal to 10. This implies a membrane that is ten times as stiff as the one in the first input file, and consequently the outputs (the z values) must all be exactly one tenth of the ones for the first input file. The testing/input.2.txt file corresponds to a 64-dimensional vector of values that are increasing in integer steps from 1 to 64. The remaining input files all have 64-dimensional vectors with random entries that were chosen as 10^x where x is a randomly distributed number between -1 and 1. In other words, the entries of the vectors stored in these remaining seven input files are all between 0.1 and 10. (Note that this choice of randomly generated numbers does not correspond to drawing from the prior probability distribution used in this program and discussed in the paper. However, for the purposes of testing, this is of course also not necessary.)
The stored output files were generated with the reference implementation of the solver by replacing the main() function with the following code:
int main()
{
const Vector<double> exact_solution(
{ 0.06076511762259369, 0.09601910120848481,
0.1238852517838584, 0.1495184117375201,
0.1841596127549784, 0.2174525028261122,
0.2250996160898698, 0.2197954769002993,
0.2074695698370926, 0.1889996477663016,
0.1632722532153726, 0.1276782480038186,
0.07711845915789312, 0.09601910120848552,
0.2000589533367983, 0.3385592591951766,
0.3934300024647806, 0.4040223892461541,
0.4122329537843092, 0.4100480091545554,
0.3949151637189968, 0.3697873264791232,
0.33401826235924, 0.2850397806663382,
0.2184260032478671, 0.1271121156350957,
0.1238852517838611, 0.3385592591951819,
0.7119285162766475, 0.8175712861756428,
0.6836254116578105, 0.5779452419831157,
0.5555615956136897, 0.5285181561736719,
0.491439702849224, 0.4409367494853282,
0.3730060082060772, 0.2821694983395214,
0.1610176733857739, 0.1495184117375257,
0.3934300024647929, 0.8175712861756562,
0.9439154625527653, 0.8015904115095128,
0.6859683749254024, 0.6561235366960599,
0.6213197201867315, 0.5753611315000049,
0.5140091754526823, 0.4325325506354165,
0.3248315148915482, 0.1834600412730086,
0.1841596127549917, 0.4040223892461832,
0.6836254116578439, 0.8015904115095396,
0.7870119561144977, 0.7373108331395808,
0.7116558878070463, 0.6745179049094283,
0.6235300574156917, 0.5559332704045935,
0.4670304994474178, 0.3499809143811,
0.19688263746294, 0.2174525028261253,
0.4122329537843404, 0.5779452419831566,
0.6859683749254372, 0.7373108331396063,
0.7458811983178246, 0.7278968022406559,
0.6904793535357751, 0.6369176452710288,
0.5677443693743215, 0.4784738764865867,
0.3602190632823262, 0.2031792054737325,
0.2250996160898818, 0.4100480091545787,
0.5555615956137137, 0.6561235366960938,
0.7116558878070715, 0.727896802240657,
0.7121928678670187, 0.6712187391428729,
0.6139157775591492, 0.5478251665295381,
0.4677122687599031, 0.3587654911000848,
0.2050734291675918, 0.2197954769003094,
0.3949151637190157, 0.5285181561736911,
0.6213197201867471, 0.6745179049094407,
0.690479353535786, 0.6712187391428787,
0.6178408289359514, 0.5453605027237883,
0.489575966490909, 0.4341716881061278,
0.3534389974779456, 0.2083227496961347,
0.207469569837099, 0.3697873264791366,
0.4914397028492412, 0.5753611315000203,
0.6235300574157017, 0.6369176452710497,
0.6139157775591579, 0.5453605027237935,
0.4336604929612851, 0.4109641743019312,
0.3881864790111245, 0.3642640090182592,
0.2179599909280145, 0.1889996477663011,
0.3340182623592461, 0.4409367494853381,
0.5140091754526943, 0.5559332704045969,
0.5677443693743304, 0.5478251665295453,
0.4895759664908982, 0.4109641743019171,
0.395727260284338, 0.3778949322004734,
0.3596268271857124, 0.2191250268948948,
0.1632722532153683, 0.2850397806663325,
0.373006008206081, 0.4325325506354207,
0.4670304994474315, 0.4784738764866023,
0.4677122687599041, 0.4341716881061055,
0.388186479011099, 0.3778949322004602,
0.3633362567187364, 0.3464457261905399,
0.2096362321365655, 0.1276782480038148,
0.2184260032478634, 0.2821694983395252,
0.3248315148915535, 0.3499809143811097,
0.3602190632823333, 0.3587654911000799,
0.3534389974779268, 0.3642640090182283,
0.35962682718569, 0.3464457261905295,
0.3260728953424643, 0.180670595355394,
0.07711845915789244, 0.1271121156350963,
0.1610176733857757, 0.1834600412730144,
0.1968826374629443, 0.2031792054737354,
0.2050734291675885, 0.2083227496961245,
0.2179599909279998, 0.2191250268948822,
0.2096362321365551, 0.1806705953553887,
0.1067965550010013 });
ForwardSimulator::PoissonSolver<2> laplace_problem(
/* global_refinements = */ 5,
/* fe_degree = */ 1,
"");
LogLikelihood::Gaussian log_likelihood(exact_solution, 0.05);
LogPrior::LogGaussian log_prior(0, 2);
const unsigned int n_theta = 64;
for (unsigned int test=0; test<10; ++test)
{
std::cout << "Generating output for test " << test << std::endl;
// For each test, read the input file...
std::ifstream test_input ("../testing/input." + std::to_string(test) + ".txt");
Assert (test_input, ExcIO());
Vector<double> theta(n_theta);
for (unsigned int i=0; i<n_theta; ++i)
test_input >> theta[i];
// ...run it through the forward simulator to get the
// z vector (which we then output to a file)...
const Vector<double> z = laplace_problem.evaluate(theta);
std::ofstream test_output_z ("output." + std::to_string(test) + ".z.txt");
z.print(test_output_z, 16);
// ...and then also evaluate prior and likelihood for these
// theta vectors:
std::ofstream test_output_likelihood ("output." + std::to_string(test) + ".likelihood.txt");
test_output_likelihood.precision(12);
test_output_likelihood << log_likelihood.log_likelihood(z) << std::endl;
std::ofstream test_output_prior ("output." + std::to_string(test) + ".prior.txt");
test_output_prior.precision(12);
test_output_prior << log_prior.log_prior(theta) << std::endl;
}
}
This code reads in each of the input files (assuming that the executable is located in a build directory parallel to the testing/ directory) and outputs its results into the current directory. The inputs you get from a modified program should then be compared against the ones stored in the testing/ directory. They should match to several digits.
An alternative implementation in Matlab
To facilitate experiments, this directory also contains alternative implementations of the benchmark. The first one was written by David Aristoff in Matlab and can be found in the Matlab/ directory. As is common in Matlab programs, each function is provided in its own file. We have verified that the program generates the same results (to 12 or more digits) as the C++ program, using the tests mentioned in the previous section.
An alternative implementation in Python
Another alternative, written by Wolfgang Bangerth, can be found in the Python/ directory and uses Python3. As is common for Python programs, the whole program is provided in one file. As for the Matlab version, we have verified that the program generates the same results (to 12 or more digits) as the C++ program, using the tests mentioned in the previous section. In fact, if you just execute the program as-is, it runs diagnostics that output the errors.
This Python version is essentially a literal translation of the Matlab code. It is not nearly as efficient (taking around 5 times as much time for each function evaluation) and could probably be optimized substantially if desired. A good starting point is the insertion of the local elements into the global matrix in the line
A[np.ix_(dof,dof)] += theta_loc * A_loc
that takes up a substantial fraction of the overall run time.
Annotated version of Matlab/exact_values.m
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%% list of "exact" measurement values, z_hat %%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
z_hat = [0.06076511762259369;
0.09601910120848481;
0.1238852517838584;
0.1495184117375201;
0.1841596127549784;
0.2174525028261122;
0.2250996160898698;
0.2197954769002993;
0.2074695698370926;
0.1889996477663016;
0.1632722532153726;
0.1276782480038186;
0.07711845915789312;
0.09601910120848552;
0.2000589533367983;
0.3385592591951766;
0.3934300024647806;
0.4040223892461541;
0.4122329537843092;
0.4100480091545554;
0.3949151637189968;
0.3697873264791232;
0.33401826235924;
0.2850397806663382;
0.2184260032478671;
0.1271121156350957;
0.1238852517838611;
0.3385592591951819;
0.7119285162766475;
0.8175712861756428;
0.6836254116578105;
0.5779452419831157;
0.5555615956136897;
0.5285181561736719;
0.491439702849224;
0.4409367494853282;
0.3730060082060772;
0.2821694983395214;
0.1610176733857739;
0.1495184117375257;
0.3934300024647929;
0.8175712861756562;
0.9439154625527653;
0.8015904115095128;
0.6859683749254024;
0.6561235366960599;
0.6213197201867315;
0.5753611315000049;
0.5140091754526823;
0.4325325506354165;
0.3248315148915482;
0.1834600412730086;
0.1841596127549917;
0.4040223892461832;
0.6836254116578439;
0.8015904115095396;
0.7870119561144977;
0.7373108331395808;
0.7116558878070463;
0.6745179049094283;
0.6235300574156917;
0.5559332704045935;
0.4670304994474178;
0.3499809143811;
0.19688263746294;
0.2174525028261253;
0.4122329537843404;
0.5779452419831566;
0.6859683749254372;
0.7373108331396063;
0.7458811983178246;
0.7278968022406559;
0.6904793535357751;
0.6369176452710288;
0.5677443693743215;
0.4784738764865867;
0.3602190632823262;
0.2031792054737325;
0.2250996160898818;
0.4100480091545787;
0.5555615956137137;
0.6561235366960938;
0.7116558878070715;
0.727896802240657;
0.7121928678670187;
0.6712187391428729;
0.6139157775591492;
0.5478251665295381;
0.4677122687599031;
0.3587654911000848;
0.2050734291675918;
0.2197954769003094;
0.3949151637190157;
0.5285181561736911;
0.6213197201867471;
0.6745179049094407;
0.690479353535786;
0.6712187391428787;
0.6178408289359514;
0.5453605027237883;
0.489575966490909;
0.4341716881061278;
0.3534389974779456;
0.2083227496961347;
0.207469569837099;
0.3697873264791366;
0.4914397028492412;
0.5753611315000203;
0.6235300574157017;
0.6369176452710497;
0.6139157775591579;
0.5453605027237935;
0.4336604929612851;
0.4109641743019312;
0.3881864790111245;
0.3642640090182592;
0.2179599909280145;
0.1889996477663011;
0.3340182623592461;
0.4409367494853381;
0.5140091754526943;
0.5559332704045969;
0.5677443693743304;
0.5478251665295453;
0.4895759664908982;
0.4109641743019171;
0.395727260284338;
0.3778949322004734;
0.3596268271857124;
0.2191250268948948;
0.1632722532153683;
0.2850397806663325;
0.373006008206081;
0.4325325506354207;
0.4670304994474315;
0.4784738764866023;
0.4677122687599041;
0.4341716881061055;
0.388186479011099;
0.3778949322004602;
0.3633362567187364;
0.3464457261905399;
0.2096362321365655;
0.1276782480038148;
0.2184260032478634;
0.2821694983395252;
0.3248315148915535;
0.3499809143811097;
0.3602190632823333;
0.3587654911000799;
0.3534389974779268;
0.3642640090182283;
0.35962682718569;
0.3464457261905295;
0.3260728953424643;
0.180670595355394;
0.07711845915789244;
0.1271121156350963;
0.1610176733857757;
0.1834600412730144;
0.1968826374629443;
0.2031792054737354;
0.2050734291675885;
0.2083227496961245;
0.2179599909279998;
0.2191250268948822;
0.2096362321365551;
0.1806705953553887;
0.1067965550010013];
Annotated version of Matlab/forward_solver.m
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%% forward solver function %%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%INPUTS:
%theta = current 8x8 parameter matrix
%lbl = cell labeling function
%A_loc = matrix of local contributions to A
%Id = Identity matrix of size 128x128
%boundaries = labels of boundary cells
%b = right hand side of linear system (AU = b)
%M = measurement matrix
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%OUTPUTS:
%z = vector of measurements
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
function z = forward_solver(theta,lbl,A_loc,Id,boundaries,b,M)
%initialize matrix A for FEM linear solve, AU = b
A = zeros(33^2,33^2);
%loop over cells to build A
for i=0:31
for j=0:31 %build A by summing over contribution from each cell
%find local coefficient in 8x8 grid
theta_loc = theta(floor(i/4)+1,floor(j/4)+1);
%update A by including contribution from cell (i,j)
dof = [lbl(i,j),lbl(i,j+1),lbl(i+1,j+1),lbl(i+1,j)];
A(dof,dof) = A(dof,dof) + theta_loc*A_loc;
end
end
%enforce boundary condition
A(boundaries,:) = Id(boundaries,:);
A(:,boundaries) = Id(:,boundaries);
%sparsify A
A = sparse(A);
%solve linear equation for coefficients, U
U = A\b;
%get new z values
z = M*U;
Annotated version of Matlab/get_statistics.m
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%% compute statistics on data set %%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%INPUTS:
%data = tensor of theta samples from each lag time and chain
%theta_means = means of theta over each independent chain
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%OUTPUTS:
%theta_mean = overall mean of chains
%covars = covariance matrices of each independent chain
%autocovar = mean of autocovariance matrix over all the chains
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
function [theta_mean,covars,autocovar] = get_statistics(data,theta_means);
%compute overall mean of data, and get size of data matrix
theta_mean = mean(theta_means,3);
[~,~,L,N] = size(data);
%initialize covariance matrices and mean autocovariance matrix
covars = zeros(64,64,N);
autocovar = zeros(64,64,2*L-1);
%compute covariance matrices and mean autocovariance matrix
for n=1:N %loop over independent Markov chains
%get data from chain n
data_ = reshape(permute(data(:,:,:,n),[3 2 1]),[L 64]);
%compute autocovariance matrix of chain n
mat = xcov(data_,'unbiased');
%store covariance matrix of chain n
covars(:,:,n) = reshape(mat(L,:),64,64);
%update mean autocovariance matrix
autocovar = autocovar + reshape(mat',[64 64 2*L-1]);
end
%compute mean of autocovariance matrix
autocovar = autocovar(1:64,1:64,L:2*L-1)/N;
Annotated version of Matlab/log_probability.m
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%% compute log probability, log pi %%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%INPUTS:
%theta = current 8x8 parameter matrix
%z = current vector of measurements
%z_hat = vector of "exact" measurements
%sig = standard deviation parameter in likelihood
%sig_pr = standard deviation parameter in prior
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%OUTPUTS:
%log_pi = logarithm of posterior probability
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
function log_pi = log_probability(theta,z,z_hat,sig,sig_pr)
%compute log likelihood
log_L = -sum((z-z_hat).^2)/(2*sig^2);
%compute log prior
log_pi_pr = -sum(log(theta).^2,'all')/(2*sig_pr^2);
%compute log posterior
log_pi = log_L+log_pi_pr;
Annotated version of Matlab/main.m
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%% Run MCMC sampler to estimate posterior distribution %%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%define number of chains, chain length, and lag time
N = input('number of independent Markov chains: ');
N_L = input('length of each Markov chain: ');
lag = input('lag time for measurements: ');
workers = input('number of parallel workers: ');
L = N_L/lag;
parpool(workers)
%load precomputations
load precomputations.mat
%define lag time and data matrix
data = zeros(8,8,L,N); %data matrix of samples at lag times
theta_means = zeros(8,8,N); %overall mean of theta
tic
parfor n=1:N
%set initial theta, theta mean, and z values of chain
theta = theta0;
theta_mean = 0;
z = z0;
for m=1:L
for l=1:lag
%define proposal, theta_tilde
xi = normrnd(0,sig_prop,[8 8]);
theta_tilde = theta.*exp(xi);
%compute new z values
z_tilde = forward_solver_(theta_tilde);
%compute posterior log probability of theta_tilde
log_pi_tilde = log_probability_(theta_tilde,z_tilde);
log_pi = log_probability_(theta,z);
%compute acceptance probability; accept proposal appropriately
accept = exp(log_pi_tilde-log_pi) ...
*prod(theta_tilde./theta,'all');
if rand<accept
theta = theta_tilde; %accept new theta values
z = z_tilde; %record associated measurements
end
%update mean of theta
theta_mean = theta_mean + theta;
end
%update data matrix
data(:,:,m,n) = theta;
end
%update theta means
theta_means(:,:,n) = theta_mean/N_L;
end
toc
%compute statistics on data set
[theta_mean,covars,autocovar] = get_statistics(data,theta_means);
%save data to Matlab workspace, labeled by N and N_L
save (['data_N_' num2str(N) '_N_L_ ' num2str(N_L) '.mat'])
Annotated version of Matlab/plot_solution.m
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%% plots solution, u, to Poisson equation %%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%% associated to theta and a 32x32 mesh %%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%input matrix theta for plotting (e.g., theta = theta_hat)
theta = input('choose theta for plotting u: theta = ');
%construct mass matrix, M_plot, for plotting
xsp = 0:0.02:1;
n = length(xsp);
Mp = zeros(n,n,33^2);
for k=1:33^2
c = inv_lbl(k);
for i=1:n
for j=1:n
Mp(i,j,k) = phi(xsp(i)-h*c(1),xsp(j)-h*c(2));
end
end
end
Mp = reshape(Mp,[n^2 33^2]);
%run forward solver on mean of theta
A = zeros(33^2,33^2);
for i=0:31
for j=0:31 %build A by summing over contribution from each cell
%find local coefficient in 8x8 grid
theta_loc = theta(floor(i/4)+1,floor(j/4)+1);
%update A by including contribution from cell (i,j)
dof = [lbl(i,j),lbl(i,j+1),lbl(i+1,j+1),lbl(i+1,j)];
A(dof,dof) = A(dof,dof) + theta_loc*A_loc;
end
end
%enforce boundary condition
A(boundaries,:) = Id(boundaries,:);
%sparsify A
A = sparse(A);
%solve linear equation for coefficients, U
U = A\b;
%close all current plots
close all
%plot solution
figure
zs = reshape(Mp*U,[n n]);
surf(xsp,xsp,zs)
Annotated version of Matlab/precomputations.m
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%% do all precomputations necessary for MCMC simulations %%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%define mesh width
h = 1/32;
%define characteristic function of unit square
S = @(x,y) heaviside(x).*heaviside(y) ...
.*(1-heaviside(x-h)).*(1-heaviside(y-h));
%define tent function on the domain [-h,h]x[-h,h]
phi = @(x,y) ((x+h).*(y+h).*S(x+h,y+h) + (h-x).*(h-y).*S(x,y) ...
+ (x+h).*(h-y).*S(x+h,y) + (h-x).*(y+h).*S(x,y+h))/h^2;
%define function that converts from (i,j) to dof, and its inverse
lbl = @(i,j) 33*j+i+1;
inv_lbl = @(k) [k-1-33*floor((k-1)/33),floor((k-1)/33)];
%construct measurement matrix, M
xs = 1/14:1/14:13/14; %measurement points
M = zeros(13,13,33^2);
for k=1:33^2
c = inv_lbl(k);
for i=1:13
for j=1:13
M(i,j,k) = phi(xs(i)-h*c(1),xs(j)-h*c(2));
end
end
end
M = reshape(M,[13^2 33^2]);
%construct exact coefficient matrix, theta_hat
theta_hat = ones(8,8);
theta_hat(2:3,2:3) = 0.1;
theta_hat(6:7,6:7) = 10;
%construct local overlap matrix, A_loc, and
identity matrix Id
A_loc = [2/3 -1/6 -1/3 -1/6;
-1/6 2/3 -1/6 -1/3;
-1/3 -1/6 2/3 -1/6;
-1/6 -1/3 -1/6 2/3];
Id = eye(33^2);
%locate boundary labels
boundaries = [lbl(0:1:32,0),lbl(0:1:32,32),lbl(0,1:1:31),lbl(32,1:1:31)];
%define RHS of FEM linear system, AU = b
b = ones(33^2,1)*10*h^2;
b(boundaries) = zeros(128,1); %enforce boundary conditions on b
%load exact z_hat values
exact_values
%set global parameters and functions for simulation
sig = 0.05; %likelihood standard deviation
sig_pr = 2; %prior (log) standard deviation
sig_prop = 0.0725; %proposal (log) standard deviation
theta0 = ones(8,8); %initial theta values
forward_solver_ = @(theta) ...
forward_solver(theta,lbl,A_loc,Id,boundaries,b,M);
log_probability_ = @(theta,z) log_probability(theta,z,z_hat,sig,sig_pr);
%find initial z values
z0 = forward_solver_(theta0);
save precomputations.mat
Annotated version of Python/forward_solver.py
import numpy as np
import scipy.sparse
from scipy.sparse.linalg import spsolve
import time
z_hat = np.array(
[0.06076511762259369, 0.09601910120848481,
0.1238852517838584, 0.1495184117375201,
0.1841596127549784, 0.2174525028261122,
0.2250996160898698, 0.2197954769002993,
0.2074695698370926, 0.1889996477663016,
0.1632722532153726, 0.1276782480038186,
0.07711845915789312, 0.09601910120848552,
0.2000589533367983, 0.3385592591951766,
0.3934300024647806, 0.4040223892461541,
0.4122329537843092, 0.4100480091545554,
0.3949151637189968, 0.3697873264791232,
0.33401826235924, 0.2850397806663382,
0.2184260032478671, 0.1271121156350957,
0.1238852517838611, 0.3385592591951819,
0.7119285162766475, 0.8175712861756428,
0.6836254116578105, 0.5779452419831157,
0.5555615956136897, 0.5285181561736719,
0.491439702849224, 0.4409367494853282,
0.3730060082060772, 0.2821694983395214,
0.1610176733857739, 0.1495184117375257,
0.3934300024647929, 0.8175712861756562,
0.9439154625527653, 0.8015904115095128,
0.6859683749254024, 0.6561235366960599,
0.6213197201867315, 0.5753611315000049,
0.5140091754526823, 0.4325325506354165,
0.3248315148915482, 0.1834600412730086,
0.1841596127549917, 0.4040223892461832,
0.6836254116578439, 0.8015904115095396,
0.7870119561144977, 0.7373108331395808,
0.7116558878070463, 0.6745179049094283,
0.6235300574156917, 0.5559332704045935,
0.4670304994474178, 0.3499809143811,
0.19688263746294, 0.2174525028261253,
0.4122329537843404, 0.5779452419831566,
0.6859683749254372, 0.7373108331396063,
0.7458811983178246, 0.7278968022406559,
0.6904793535357751, 0.6369176452710288,
0.5677443693743215, 0.4784738764865867,
0.3602190632823262, 0.2031792054737325,
0.2250996160898818, 0.4100480091545787,
0.5555615956137137, 0.6561235366960938,
0.7116558878070715, 0.727896802240657,
0.7121928678670187, 0.6712187391428729,
0.6139157775591492, 0.5478251665295381,
0.4677122687599031, 0.3587654911000848,
0.2050734291675918, 0.2197954769003094,
0.3949151637190157, 0.5285181561736911,
0.6213197201867471, 0.6745179049094407,
0.690479353535786, 0.6712187391428787,
0.6178408289359514, 0.5453605027237883,
0.489575966490909, 0.4341716881061278,
0.3534389974779456, 0.2083227496961347,
0.207469569837099, 0.3697873264791366,
0.4914397028492412, 0.5753611315000203,
0.6235300574157017, 0.6369176452710497,
0.6139157775591579, 0.5453605027237935,
0.4336604929612851, 0.4109641743019312,
0.3881864790111245, 0.3642640090182592,
0.2179599909280145, 0.1889996477663011,
0.3340182623592461, 0.4409367494853381,
0.5140091754526943, 0.5559332704045969,
0.5677443693743304, 0.5478251665295453,
0.4895759664908982, 0.4109641743019171,
0.395727260284338, 0.3778949322004734,
0.3596268271857124, 0.2191250268948948,
0.1632722532153683, 0.2850397806663325,
0.373006008206081, 0.4325325506354207,
0.4670304994474315, 0.4784738764866023,
0.4677122687599041, 0.4341716881061055,
0.388186479011099, 0.3778949322004602,
0.3633362567187364, 0.3464457261905399,
0.2096362321365655, 0.1276782480038148,
0.2184260032478634, 0.2821694983395252,
0.3248315148915535, 0.3499809143811097,
0.3602190632823333, 0.3587654911000799,
0.3534389974779268, 0.3642640090182283,
0.35962682718569, 0.3464457261905295,
0.3260728953424643, 0.180670595355394,
0.07711845915789244, 0.1271121156350963,
0.1610176733857757, 0.1834600412730144,
0.1968826374629443, 0.2031792054737354,
0.2050734291675885, 0.2083227496961245,
0.2179599909279998, 0.2191250268948822,
0.2096362321365551, 0.1806705953553887,
0.1067965550010013])
h = 1/32
def heaviside(x) :
if x<0 :
return 0
else :
return 1
def S(x,y) :
return heaviside(x)*heaviside(y) * (1-heaviside(x-h))*(1-heaviside(y-h));
def phi(x,y) :
return ((x+h)*(y+h)*S(x+h,y+h) + (h-x)*(h-y)*S(x,y)
+ (x+h)*(h-y)*S(x+h,y) + (h-x)*(y+h)*S(x,y+h))/h**2
def ij_to_dof_index(i,j) :
return 33*j+i
def inv_ij_to_dof_index(k) :
return [k-33*
int(k/33),
int(k/33)]
xs = np.arange(1./14,13./14,1./14);
M = np.zeros((13,13,33**2));
for k in range(33**2) :
c = inv_ij_to_dof_index(k)
for i in range(13) :
for j in range(13) :
M[i,j,k] = phi(xs[i]-h*c[0], xs[j]-h*c[1])
M = M.reshape((13**2, 33**2))
M = scipy.sparse.csr_matrix(M);
A_loc = np.array([[2./3, -1./6, -1./3, -1./6],
[-1./6, 2./3, -1./6, -1./3],
[-1./3, -1./6, 2./3, -1./6],
[-1./6, -1./3, -1./6, 2./3]])
Id = np.eye(33**2,33**2)
boundaries = ([ij_to_dof_index(i,0) for i in range(33)] +
[ij_to_dof_index(i,32) for i in range(33)] +
[ij_to_dof_index(0,j+1) for j in range(31)] +
[ij_to_dof_index(32,j+1) for j in range(31)])
b = np.ones(33**2)*10*h**2
b[boundaries] = 0
def forward_solver(theta) :
A = np.zeros((33**2,33**2))
for i in range(32) :
for j in range (32) :
theta_loc = theta[
int(i/4)+
int(j/4)*8]
dof = [ij_to_dof_index(i,j),
ij_to_dof_index(i,j+1),
ij_to_dof_index(i+1,j+1),
ij_to_dof_index(i+1,j)]
A[np.ix_(dof,dof)] += theta_loc * A_loc
A[boundaries,:] = 0
A[:,boundaries] = 0
A[boundaries,boundaries] = 1
u = spsolve(scipy.sparse.csr_matrix(A), b)
z = M * u
return z
def log_likelihood(theta) :
z = forward_solver(theta)
misfit = z - z_hat
sig = 0.05
return -np.dot(misfit,misfit)/(2*sig**2)
def log_prior(theta) :
sig_pr = 2
return -np.linalg.norm(np.log(theta))**2/(2*sig_pr**2)
def log_posterior(theta) :
return log_likelihood(theta) + log_prior(theta)
def verify_against_stored_tests() :
for i in range(10) :
print ("Verifying against data set", i)
f_input = open ("../testing/input.{}.txt".format(i), 'r')
theta = np.fromfile(f_input, count=64, sep=" ")
this_z = forward_solver(theta)
this_log_likelihood = log_likelihood(theta)
this_log_prior = log_prior(theta)
f_output_z = open ("../testing/output.{}.z.txt".format(i), 'r')
f_output_likelihood = open ("../testing/output.{}.likelihood.txt".format(i), 'r')
f_output_prior = open ("../testing/output.{}.prior.txt".format(i), 'r')
reference_z = np.fromfile(f_output_z, count=13**2, sep=" ")
reference_log_likelihood = float(f_output_likelihood.read())
reference_log_prior = float(f_output_prior.read())
print (" || z-z_ref || : ",
np.linalg.norm(this_z - reference_z))
print (" log likelihood : ",
"Python value=", this_log_likelihood,
"(C++ reference value=", reference_log_likelihood,
", error=", abs(this_log_likelihood - reference_log_likelihood),
")")
print (" log prior : ",
"Python value=", this_log_prior,
"(C++ reference value=", reference_log_prior,
", error=", abs(this_log_prior - reference_log_prior),
")")
def time_forward_solver() :
begin = time.time()
n_runs = 100
for i in range(n_runs) :
theta = np.exp(np.random.rand(64) * 4 - 2)
z = forward_solver(theta)
end = time.time()
print ("Time per forward evaluation:", (end-begin)/n_runs)
verify_against_stored_tests()
time_forward_solver()
Annotated version of mcmc-laplace.cc
#include <fstream>
#include <iostream>
#include <random>
The following is a namespace in which we define the solver of the PDE. The main class implements an abstract Interface class declared at the top, which provides for an evaluate() function that, given a coefficient vector, solves the PDE discussed in the Readme file and then evaluates the solution at the 169 mentioned points.
The solver follows the basic layout of step-4, though it precomputes a number of things in the setup_system() function, such as the evaluation of the matrix that corresponds to the point evaluations, as well as the local contributions to matrix and right hand side.
Rather than commenting on everything in detail, in the following we will only document those things that are not already clear from step-4 and a small number of other tutorial programs.
namespace ForwardSimulator
{
class Interface
{
public:
virtual ~Interface() = default;
};
template <int dim>
class PoissonSolver : public Interface
{
public:
PoissonSolver(const unsigned int global_refinements,
const unsigned int fe_degree,
const std::string &dataset_name);
private:
void make_grid(const unsigned int global_refinements);
void setup_system();
void solve();
std::map<types::global_dof_index,double> boundary_values;
std::vector<Point<dim>> measurement_points;
unsigned int nth_evaluation;
const std::string &dataset_name;
};
template <int dim>
PoissonSolver<dim>::PoissonSolver(const unsigned int global_refinements,
const unsigned int fe_degree,
const std::string &dataset_name)
: fe(fe_degree)
, nth_evaluation(0)
, dataset_name(dataset_name)
{
make_grid(global_refinements);
setup_system();
}
template <int dim>
void PoissonSolver<dim>::make_grid(const unsigned int global_refinements)
{
Assert(global_refinements >= 3,
ExcMessage("This program makes the assumption that the mesh for the "
"solution of the PDE is at least as fine as the one used "
"in the definition of the coefficient."));
std::cout <<
" Number of active cells: " <<
triangulation.n_active_cells()
<< std::endl;
}
template <int dim>
void PoissonSolver<dim>::setup_system()
{
#define Assert(cond, exc)
void hyper_cube(Triangulation< dim, spacedim > &tria, const double left=0., const double right=1., const bool colorize=false)
const ::parallel::distributed::Triangulation< dim, spacedim > * triangulation
First define the finite element space:
dof_handler.distribute_dofs(fe);
std::cout << " Number of degrees of freedom: " << dof_handler.n_dofs()
<< std::endl;
Then set up the main data structures that will hold the discrete problem:
{
sparsity_pattern.copy_from(dsp);
system_matrix.reinit(sparsity_pattern);
solution.reinit(dof_handler.n_dofs());
system_rhs.reinit(dof_handler.n_dofs());
}
void make_sparsity_pattern(const DoFHandler< dim, spacedim > &dof_handler, SparsityPatternType &sparsity_pattern, const AffineConstraints< number > &constraints=AffineConstraints< number >(), const bool keep_constrained_dofs=true, const types::subdomain_id subdomain_id=numbers::invalid_subdomain_id)
And then define the tools to do point evaluation. We choose a set of 13x13 points evenly distributed across the domain:
{
const unsigned int n_points_per_direction = 13;
const double dx = 1. / (n_points_per_direction + 1);
for (unsigned int x = 1; x <= n_points_per_direction; ++x)
for (unsigned int y = 1; y <= n_points_per_direction; ++y)
measurement_points.emplace_back(x * dx, y * dx);
First build a full matrix of the evaluation process. We do this even though the matrix is really sparse – but we don't know which entries are nonzero. Later, the copy_from() function calls build a sparsity pattern and a sparse matrix from the dense matrix.
n_points_per_direction,
dof_handler.n_dofs());
for (unsigned int index = 0; index < measurement_points.size(); ++index)
{
measurement_points[index],
weights);
for (unsigned int i = 0; i < dof_handler.n_dofs(); ++i)
full_measurement_matrix(index, i) = weights(i);
}
measurement_sparsity.copy_from(full_measurement_matrix);
measurement_matrix.reinit(measurement_sparsity);
measurement_matrix.copy_from(full_measurement_matrix);
}
Next build the mapping from cell to the index in the 64-element coefficient vector:
{
const unsigned int i = std::floor(cell->center()[0] * 8);
const unsigned int j = std::floor(cell->center()[1] * 8);
const unsigned int index = i + 8 * j;
cell->set_user_index(index);
}
Finally prebuild the building blocks of the linear system as discussed in the Readme file :
{
const unsigned int dofs_per_cell = fe.dofs_per_cell;
cell_matrix.reinit(dofs_per_cell, dofs_per_cell);
cell_rhs.reinit(dofs_per_cell);
const unsigned int n_q_points = quadrature_formula.size();
quadrature_formula,
fe_values.reinit(dof_handler.begin_active());
for (unsigned int q_index = 0; q_index < n_q_points; ++q_index)
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
for (unsigned int j = 0; j < dofs_per_cell; ++j)
cell_matrix(i, j) +=
(fe_values.shape_grad(i, q_index) *
fe_values.shape_grad(j, q_index) *
fe_values.JxW(q_index));
cell_rhs(i) += (fe_values.shape_value(i, q_index) *
10.0 *
fe_values.JxW(q_index));
}
0,
boundary_values);
}
}
@ update_values
Shape function values.
@ update_JxW_values
Transformed quadrature weights.
@ update_gradients
Shape function gradients.
Given that we have pre-built the matrix and right hand side contributions for a (representative) cell, the function that assembles the matrix is pretty short and straightforward:
template <int dim>
void PoissonSolver<dim>::assemble_system(
const Vector<double> &coefficients)
{
Assert(coefficients.
size() == 64, ExcInternalError());
system_matrix = 0;
system_rhs = 0;
const unsigned int dofs_per_cell = fe.dofs_per_cell;
std::vector<types::global_dof_index> local_dof_indices(dofs_per_cell);
for (const auto &cell : dof_handler.active_cell_iterators())
{
const double coefficient = coefficients(cell->user_index());
cell->get_dof_indices(local_dof_indices);
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
for (unsigned int j = 0; j < dofs_per_cell; ++j)
system_matrix.add(local_dof_indices[i],
local_dof_indices[j],
coefficient * cell_matrix(i, j));
system_rhs(local_dof_indices[i]) += cell_rhs(i);
}
}
system_matrix,
solution,
system_rhs);
}
The same is true for the function that solves the linear system:
template <int dim>
void PoissonSolver<dim>::solve()
{
solver.solve(system_matrix, solution, system_rhs, ilu);
}
void initialize(const SparseMatrix< somenumber > &matrix, const AdditionalData ¶meters=AdditionalData())
The following function outputs graphical data for the most recently used coefficient and corresponding solution of the PDE. Collecting the coefficient values requires translating from the 64-element coefficient vector and the cells that correspond to each of these elements. The rest remains pretty obvious, with the exception of including the number of the current sample into the file name.
template <int dim>
void
PoissonSolver<dim>::output_results(
const Vector<double> &coefficients)
const{
coefficient_values[cell->active_cell_index()] =
coefficients(cell->user_index());
std::ofstream output("solution-" +
}
void attach_dof_handler(const DoFHandler< dim, spacedim > &)
void add_data_vector(const VectorType &data, const std::vector< std::string > &names, const DataVectorType type=type_automatic, const std::vector< DataComponentInterpretation::DataComponentInterpretation > &data_component_interpretation={})
virtual void build_patches(const unsigned int n_subdivisions=0)
void write_vtu(std::ostream &out) const
std::string int_to_string(const unsigned int value, const unsigned int digits=numbers::invalid_unsigned_int)
The following is the main function of this class: Given a coefficient vector, it assembles the linear system, solves it, and then evaluates the solution at the measurement points by applying the measurement matrix to the solution vector. That vector of "measured" values is then returned.
The function will also output the solution in a graphical format if you un-comment the corresponding statement in the third code block. However, you may end up with a very large amount of data: This code is producing, at the minimum, 10,000 samples and creating output for each one of them is surely more data than you ever want to see!
At the end of the function, we output some timing information every 10,000 samples.
template <int dim>
{
{
assemble_system(coefficients);
}
{
solve();
}
{
measurement_matrix.vmult(measurements, solution);
Assert(measurements.size() == measurement_points.size(),
ExcInternalError());
}
++nth_evaluation;
if (nth_evaluation % 10000 == 0)
timer.print_summary();
return measurements;
}
}
The following namespaces define the statistical properties of the Bayesian inverse problem. The first is about the definition of the measurement statistics (the "likelihood"), which we here assume to be a normal distribution \(N(\mu,\sigma I)\) with mean value \(\mu\) given by the actual measurement vector (passed as an argument to the constructor of the Gaussian class and standard deviation \(\sigma\).
For reasons of numerical accuracy, it is useful to not return the actual likelihood, but its logarithm. This is because these values can be very small, occasionally on the order of \(e^{-100}\), for which it becomes very difficult to compute accurate values.
namespace LogLikelihood
{
class Interface
{
public:
virtual ~Interface() = default;
};
class Gaussian : public Interface
{
public:
virtual double log_likelihood(
const Vector<double> &x)
const override;
private:
const double sigma;
};
: mu(mu)
, sigma(sigma)
{}
{
x_minus_mu -= mu;
return -x_minus_mu.
norm_sqr() / (2 * sigma * sigma);
}
}
real_type norm_sqr() const
Next up is the "prior" imposed on the coefficients. We assume that the logarithms of the entries of the coefficient vector are all distributed as a Gaussian with given mean and standard deviation. If the logarithms of the coefficients are normally distributed, then this implies in particular that the coefficients can only be positive, which is a useful property to ensure the well-posedness of the forward problem.
For the same reasons as for the likelihood above, the interface for the prior asks for returning the logarithm of the prior, instead of the prior probability itself.
namespace LogPrior
{
class Interface
{
public:
virtual ~Interface() = default;
};
class LogGaussian : public Interface
{
public:
LogGaussian(const double mu, const double sigma);
private:
const double mu;
const double sigma;
};
LogGaussian::LogGaussian(const double mu, const double sigma)
: mu(mu)
, sigma(sigma)
{}
{
double log_of_product = 0;
for (const auto &el : x)
log_of_product +=
return log_of_product;
}
}
::VectorizedArray< Number, width > log(const ::VectorizedArray< Number, width > &)
The Metropolis-Hastings algorithm requires a method to create a new sample given a previous sample. We do this by perturbing the current (coefficient) sample randomly using a Gaussian distribution centered at the current sample. To ensure that the samples' individual entries all remain positive, we use a Gaussian distribution in logarithm space – in other words, instead of adding a small perturbation with mean value zero, we multiply the entries of the current sample by a factor that is the exponential of a random number with mean zero. (Because the exponential of zero is one, this means that the most likely factors to multiply the existing sample entries by are close to one. And because the exponential of a number is always positive, we never get negative samples this way.)
But the Metropolis-Hastings sampler doesn't just need a perturbed sample \(y\) location given the current sample location \(x\). It also needs to know the ratio of the probability of reaching \(y\) from \(x\), divided by the probability of reaching \(x\) from \(y\). If we were to use a symmetric proposal distribution (e.g., a Gaussian distribution centered at \(x\) with a width independent of \(x\)), then these two probabilities would be the same, and the ratio one. But that's not the case for the Gaussian in log space. It's not terribly difficult to verify that in that case, for a single component the ratio of these probabilities is \(y_i/x_i\), and consequently for all components of the vector together, the probability is the product of these ratios.
namespace ProposalGenerator
{
class Interface
{
public:
virtual
std::pair<Vector<double>,double>
virtual ~Interface() = default;
};
class LogGaussian : public Interface
{
public:
LogGaussian(const unsigned int random_seed, const double log_sigma);
virtual
std::pair<Vector<double>,double>
private:
const double log_sigma;
mutable std::mt19937 random_number_generator;
};
LogGaussian::LogGaussian(const unsigned int random_seed,
const double log_sigma)
: log_sigma(log_sigma)
{
random_number_generator.seed(random_seed);
}
std::pair<Vector<double>,double>
{
double product_of_ratios = 1;
for (auto &x : new_sample)
{
const double rnd = std::normal_distribution<>(0, log_sigma)(random_number_generator);
x *= exp_rnd;
product_of_ratios *= exp_rnd;
}
return {new_sample, product_of_ratios};
}
}
::VectorizedArray< Number, width > exp(const ::VectorizedArray< Number, width > &)
The last main class is the Metropolis-Hastings sampler itself. If you understand the algorithm behind this method, then the following implementation should not be too difficult to read. The only thing of relevance is that descriptions of the algorithm typically ask whether the ratio of two probabilities (the "posterior" probabilities of the current and the previous samples, where the "posterior" is the product of the likelihood and the prior probability) is larger or smaller than a randomly drawn number. But because our interfaces return the logarithms of these probabilities, we now need to take the ratio of appropriate exponentials – which is made numerically more stable by considering the exponential of the difference of the log probabilities. The only other slight complication is that we need to multiply this ratio by the ratio of proposal probabilities since we use a non-symmetric proposal distribution.
Finally, we note that the output is generated with 7 digits of accuracy. (The C++ default is 6 digits.) We do this because, as shown in the paper, we can determine the mean value of the probability distribution we are sampling here to at least six digits of accuracy, and do not want to be limited by the precision of the output.
namespace Sampler
{
class MetropolisHastings
{
public:
MetropolisHastings(ForwardSimulator::Interface & simulator,
const LogLikelihood::Interface & likelihood,
const LogPrior::Interface & prior,
const ProposalGenerator::Interface &proposal_generator,
const unsigned int random_seed,
const std::string & dataset_name);
const unsigned int n_samples);
private:
ForwardSimulator::Interface & simulator;
const LogLikelihood::Interface & likelihood;
const LogPrior::Interface & prior;
const ProposalGenerator::Interface &proposal_generator;
std::mt19937 random_number_generator;
unsigned int sample_number;
unsigned int accepted_sample_number;
std::ofstream output_file;
const double current_log_likelihood);
};
MetropolisHastings::MetropolisHastings(
ForwardSimulator::Interface & simulator,
const LogLikelihood::Interface & likelihood,
const LogPrior::Interface & prior,
const ProposalGenerator::Interface &proposal_generator,
const unsigned int random_seed,
const std::string & dataset_name)
: simulator(simulator)
, likelihood(likelihood)
, prior(prior)
, proposal_generator(proposal_generator)
, sample_number(0)
, accepted_sample_number(0)
{
output_file.open("samples-" + dataset_name + ".txt");
output_file.precision(7);
random_number_generator.seed(random_seed);
}
const unsigned int n_samples)
{
std::uniform_real_distribution<> uniform_distribution(0, 1);
double current_log_posterior =
(likelihood.log_likelihood(simulator.evaluate(current_sample)) +
prior.log_prior(current_sample));
++sample_number;
++accepted_sample_number;
write_sample(current_sample, current_log_posterior);
for (unsigned int k = 1; k < n_samples; ++k, ++sample_number)
{
std::pair<Vector<double>,double>
perturbation = proposal_generator.perturb(current_sample);
const double perturbation_probability_ratio = perturbation.second;
const double trial_log_posterior =
(likelihood.log_likelihood(simulator.evaluate(trial_sample)) +
prior.log_prior(trial_sample));
if (
std::exp(trial_log_posterior - current_log_posterior) * perturbation_probability_ratio
>=
uniform_distribution(random_number_generator))
{
current_sample = trial_sample;
current_log_posterior = trial_log_posterior;
++accepted_sample_number;
}
write_sample(current_sample, current_log_posterior);
}
}
void MetropolisHastings::write_sample(
const Vector<double> ¤t_sample,
const double current_log_posterior)
{
output_file << current_log_posterior << '\t';
output_file << accepted_sample_number << '\t';
for (const auto &x : current_sample)
output_file << x << ' ';
output_file << '\n';
output_file.flush();
}
}
The final function is main(), which simply puts all of these pieces together into one. The "exact solution", i.e., the "measurement values" we use for this program are tabulated to make it easier for other people to use in their own implementations of this benchmark. These values created using the same main class above, but using 8 mesh refinements and using a Q3 element – i.e., using a much more accurate method than the one we use in the forward simulator for generating samples below (which uses 5 global mesh refinement steps and a Q1 element). If you wanted to regenerate this set of numbers, then the following code snippet would do that:
int main()
{
const bool testing = true;
Run with one thread, so as to not step on other processes doing the same at the same time. It turns out that the problem is also so small that running with more than one thread increases the runtime.
const unsigned int random_seed = (testing ? 1U : std::random_device()());
{ 0.06076511762259369, 0.09601910120848481,
0.1238852517838584, 0.1495184117375201,
0.1841596127549784, 0.2174525028261122,
0.2250996160898698, 0.2197954769002993,
0.2074695698370926, 0.1889996477663016,
0.1632722532153726, 0.1276782480038186,
0.07711845915789312, 0.09601910120848552,
0.2000589533367983, 0.3385592591951766,
0.3934300024647806, 0.4040223892461541,
0.4122329537843092, 0.4100480091545554,
0.3949151637189968, 0.3697873264791232,
0.33401826235924, 0.2850397806663382,
0.2184260032478671, 0.1271121156350957,
0.1238852517838611, 0.3385592591951819,
0.7119285162766475, 0.8175712861756428,
0.6836254116578105, 0.5779452419831157,
0.5555615956136897, 0.5285181561736719,
0.491439702849224, 0.4409367494853282,
0.3730060082060772, 0.2821694983395214,
0.1610176733857739, 0.1495184117375257,
0.3934300024647929, 0.8175712861756562,
0.9439154625527653, 0.8015904115095128,
0.6859683749254024, 0.6561235366960599,
0.6213197201867315, 0.5753611315000049,
0.5140091754526823, 0.4325325506354165,
0.3248315148915482, 0.1834600412730086,
0.1841596127549917, 0.4040223892461832,
0.6836254116578439, 0.8015904115095396,
0.7870119561144977, 0.7373108331395808,
0.7116558878070463, 0.6745179049094283,
0.6235300574156917, 0.5559332704045935,
0.4670304994474178, 0.3499809143811,
0.19688263746294, 0.2174525028261253,
0.4122329537843404, 0.5779452419831566,
0.6859683749254372, 0.7373108331396063,
0.7458811983178246, 0.7278968022406559,
0.6904793535357751, 0.6369176452710288,
0.5677443693743215, 0.4784738764865867,
0.3602190632823262, 0.2031792054737325,
0.2250996160898818, 0.4100480091545787,
0.5555615956137137, 0.6561235366960938,
0.7116558878070715, 0.727896802240657,
0.7121928678670187, 0.6712187391428729,
0.6139157775591492, 0.5478251665295381,
0.4677122687599031, 0.3587654911000848,
0.2050734291675918, 0.2197954769003094,
0.3949151637190157, 0.5285181561736911,
0.6213197201867471, 0.6745179049094407,
0.690479353535786, 0.6712187391428787,
0.6178408289359514, 0.5453605027237883,
0.489575966490909, 0.4341716881061278,
0.3534389974779456, 0.2083227496961347,
0.207469569837099, 0.3697873264791366,
0.4914397028492412, 0.5753611315000203,
0.6235300574157017, 0.6369176452710497,
0.6139157775591579, 0.5453605027237935,
0.4336604929612851, 0.4109641743019312,
0.3881864790111245, 0.3642640090182592,
0.2179599909280145, 0.1889996477663011,
0.3340182623592461, 0.4409367494853381,
0.5140091754526943, 0.5559332704045969,
0.5677443693743304, 0.5478251665295453,
0.4895759664908982, 0.4109641743019171,
0.395727260284338, 0.3778949322004734,
0.3596268271857124, 0.2191250268948948,
0.1632722532153683, 0.2850397806663325,
0.373006008206081, 0.4325325506354207,
0.4670304994474315, 0.4784738764866023,
0.4677122687599041, 0.4341716881061055,
0.388186479011099, 0.3778949322004602,
0.3633362567187364, 0.3464457261905399,
0.2096362321365655, 0.1276782480038148,
0.2184260032478634, 0.2821694983395252,
0.3248315148915535, 0.3499809143811097,
0.3602190632823333, 0.3587654911000799,
0.3534389974779268, 0.3642640090182283,
0.35962682718569, 0.3464457261905295,
0.3260728953424643, 0.180670595355394,
0.07711845915789244, 0.1271121156350963,
0.1610176733857757, 0.1834600412730144,
0.1968826374629443, 0.2031792054737354,
0.2050734291675885, 0.2083227496961245,
0.2179599909279998, 0.2191250268948822,
0.2096362321365551, 0.1806705953553887,
0.1067965550010013 });
static void set_thread_limit(const unsigned int max_threads=numbers::invalid_unsigned_int)
std::string to_string(const number value, const unsigned int digits=numbers::invalid_unsigned_int)
Now run the forward simulator for samples:
ForwardSimulator::PoissonSolver<2> laplace_problem(
5,
1,
dataset_name);
LogLikelihood::Gaussian log_likelihood(exact_solution, 0.05);
LogPrior::LogGaussian log_prior(0, 2);
ProposalGenerator::LogGaussian proposal_generator(
random_seed, 0.09);
Sampler::MetropolisHastings sampler(laplace_problem,
log_likelihood,
log_prior,
proposal_generator,
random_seed,
dataset_name);
for (auto &el : starting_coefficients)
el = 1.;
sampler.sample(starting_coefficients,
(testing ? 250 * 40
:
100000000
));
}




 The coefficient values that correspond to this solution (the "exact" coefficient from which the measurements \(\hat{\mathbf z}\) were generated) looks as follows:
The coefficient values that correspond to this solution (the "exact" coefficient from which the measurements \(\hat{\mathbf z}\) were generated) looks as follows: 