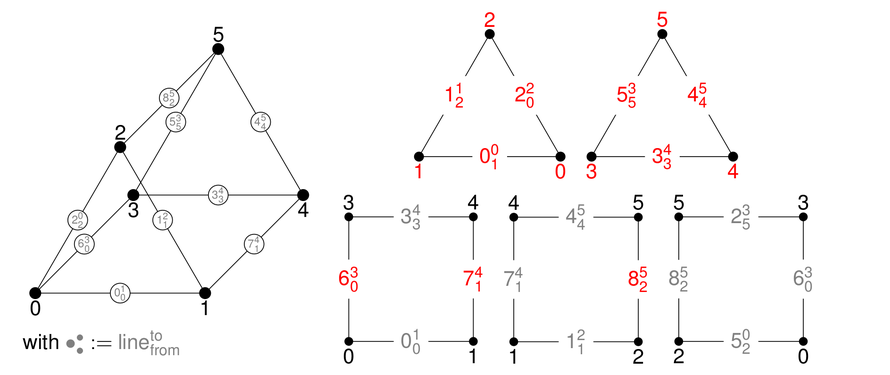
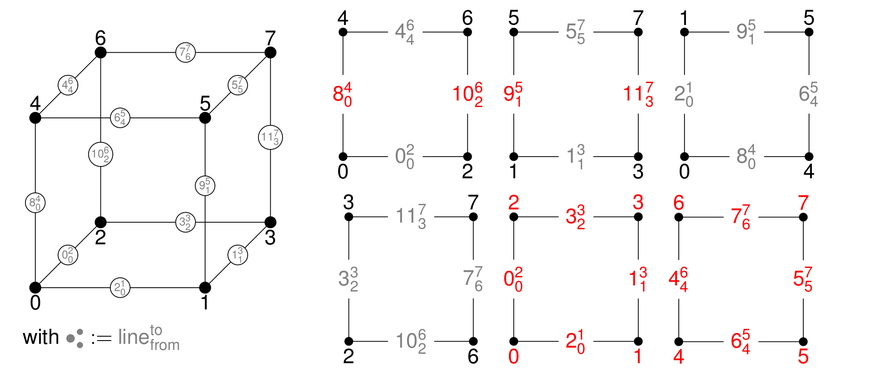
This module describes the experimental simplex support in deal.II.
More...
|
| class | ScalarLagrangePolynomialPyramid< dim > |
| |
| class | QGaussSimplex< dim > |
| |
| class | QWitherdenVincentSimplex< dim > |
| |
| class | FE_PyramidPoly< dim, spacedim > |
| |
| class | FE_PyramidP< dim, spacedim > |
| |
| class | FE_PyramidDGP< dim, spacedim > |
| |
| class | FE_SimplexPoly< dim, spacedim > |
| |
| class | FE_SimplexP< dim, spacedim > |
| |
| class | FE_SimplexDGP< dim, spacedim > |
| |
| class | FE_WedgePoly< dim, spacedim > |
| |
| class | FE_WedgeP< dim, spacedim > |
| |
| class | FE_WedgeDGP< dim, spacedim > |
| |
| class | MappingFE< dim, spacedim > |
| |
|
| template<int dim, int spacedim> |
| void | GridGenerator::subdivided_hyper_rectangle_with_simplices (Triangulation< dim, spacedim > &tria, const std::vector< unsigned int > &repetitions, const Point< dim > &p1, const Point< dim > &p2, const bool colorize=false) |
| |
| template<int dim, int spacedim> |
| void | GridGenerator::subdivided_hyper_cube_with_simplices (Triangulation< dim, spacedim > &tria, const unsigned int repetitions, const double p1=0.0, const double p2=1.0, const bool colorize=false) |
| |
This module describes the experimental simplex support in deal.II.
Simplex and mixed meshes in deal.II are still experimental, i.e., work in progress. Large parts of the library have been ported to be able to operate on such kind of meshes. However, there are still many functions that need to be generalized. You can get a good overview of the ported functionalities by taking a look at the tests in the folder "tests/simplex". In the following, we provide two very basic examples to get started and provide some implementation details.
Example: simplex mesh
The following code shows how to work with simplex meshes:
#include <fstream>
#include <iostream>
class Step3
{
public:
Step3();
private:
void make_grid();
void setup_system();
void assemble_system();
void solve();
void output_results() const;
};
Step3::Step3()
, fe(2)
, quadrature_formula(3)
{}
void Step3::make_grid()
{
std::cout <<
"Number of active cells: " <<
triangulation.n_active_cells()
<< std::endl;
}
void Step3::setup_system()
{
dof_handler.distribute_dofs(fe);
std::cout << "Number of degrees of freedom: " << dof_handler.n_dofs()
<< std::endl;
sparsity_pattern.copy_from(dsp);
system_matrix.reinit(sparsity_pattern);
solution.reinit(dof_handler.n_dofs());
system_rhs.reinit(dof_handler.n_dofs());
}
void Step3::assemble_system()
{
fe,
quadrature_formula,
const unsigned int dofs_per_cell = fe.n_dofs_per_cell();
std::vector<types::global_dof_index> local_dof_indices(dofs_per_cell);
for (const auto &cell : dof_handler.active_cell_iterators())
{
fe_values.reinit(cell);
cell_rhs = 0;
for (const unsigned int q_index : fe_values.quadrature_point_indices())
{
for (const unsigned int i : fe_values.dof_indices())
for (const unsigned int j : fe_values.dof_indices())
(fe_values.shape_grad(i, q_index) *
fe_values.shape_grad(j, q_index) *
fe_values.JxW(q_index));
for (const unsigned int i : fe_values.dof_indices())
cell_rhs(i) += (fe_values.shape_value(i, q_index) *
1. *
fe_values.JxW(q_index));
}
cell->get_dof_indices(local_dof_indices);
for (const unsigned int i : fe_values.dof_indices())
for (const unsigned int j : fe_values.dof_indices())
system_matrix.add(local_dof_indices[i],
local_dof_indices[j],
for (const unsigned int i : fe_values.dof_indices())
system_rhs(local_dof_indices[i]) += cell_rhs(i);
}
std::map<types::global_dof_index, double> boundary_values;
system_matrix,
solution,
system_rhs);
}
void Step3::solve()
{
}
void Step3::output_results() const
{
std::ofstream output("solution.vtk");
}
{
make_grid();
setup_system();
assemble_system();
solve();
output_results();
}
int main()
{
Step3 laplace_problem;
laplace_problem.run();
return 0;
}
void attach_dof_handler(const DoFHandlerType &)
void add_data_vector(const VectorType &data, const std::vector< std::string > &names, const DataVectorType type=type_automatic, const std::vector< DataComponentInterpretation::DataComponentInterpretation > &data_component_interpretation=std::vector< DataComponentInterpretation::DataComponentInterpretation >())
virtual void build_patches(const unsigned int n_subdivisions=0)
void read(std::istream &in, Format format=Default)
unsigned int depth_console(const unsigned int n)
@ update_values
Shape function values.
@ update_JxW_values
Transformed quadrature weights.
@ update_gradients
Shape function gradients.
bool write_higher_order_cells
void set_flags(const FlagType &flags)
void write_vtk(std::ostream &out) const
void make_sparsity_pattern(const DoFHandler< dim, spacedim > &dof_handler, SparsityPatternType &sparsity_pattern, const AffineConstraints< number > &constraints=AffineConstraints< number >(), const bool keep_constrained_dofs=true, const types::subdomain_id subdomain_id=numbers::invalid_subdomain_id)
void cell_matrix(FullMatrix< double > &M, const FEValuesBase< dim > &fe, const FEValuesBase< dim > &fetest, const ArrayView< const std::vector< double > > &velocity, const double factor=1.)
SymmetricTensor< 2, dim, Number > e(const Tensor< 2, dim, Number > &F)
void run(const Iterator &begin, const typename identity< Iterator >::type &end, Worker worker, Copier copier, const ScratchData &sample_scratch_data, const CopyData &sample_copy_data, const unsigned int queue_length, const unsigned int chunk_size)
const ::parallel::distributed::Triangulation< dim, spacedim > * triangulation
Example: mixed mesh
The following code shows how to work with mixed meshes:
#include <fstream>
#include <iostream>
class Step3
{
public:
Step3();
private:
void make_grid();
void setup_system();
void assemble_system();
void solve();
void output_results() const;
};
Step3::Step3()
{}
void Step3::make_grid()
{
std::cout <<
"Number of active cells: " <<
triangulation.n_active_cells()
<< std::endl;
}
void Step3::setup_system()
{
for (const auto &cell : dof_handler.active_cell_iterators())
{
cell->set_active_fe_index(0);
cell->set_active_fe_index(1);
else
}
dof_handler.distribute_dofs(fe);
std::cout << "Number of degrees of freedom: " << dof_handler.n_dofs()
<< std::endl;
sparsity_pattern.copy_from(dsp);
system_matrix.reinit(sparsity_pattern);
solution.reinit(dof_handler.n_dofs());
system_rhs.reinit(dof_handler.n_dofs());
}
void Step3::assemble_system()
{
fe,
quadrature_formula,
std::vector<types::global_dof_index> local_dof_indices;
for (const auto &cell : dof_handler.active_cell_iterators())
{
const auto &fe_values = hp_fe_values.get_present_fe_values();
const unsigned int dofs_per_cell = cell->get_fe().n_dofs_per_cell();
cell_rhs.
reinit(dofs_per_cell);
local_dof_indices.resize(dofs_per_cell);
cell_rhs = 0;
for (const unsigned int q_index : fe_values.quadrature_point_indices())
{
for (const unsigned int i : fe_values.dof_indices())
for (const unsigned int j : fe_values.dof_indices())
(fe_values.shape_grad(i, q_index) *
fe_values.shape_grad(j, q_index) *
fe_values.JxW(q_index));
for (const unsigned int i : fe_values.dof_indices())
cell_rhs(i) += (fe_values.shape_value(i, q_index) *
1. *
fe_values.JxW(q_index));
}
cell->get_dof_indices(local_dof_indices);
for (const unsigned int i : fe_values.dof_indices())
for (const unsigned int j : fe_values.dof_indices())
system_matrix.
add(local_dof_indices[i],
local_dof_indices[j],
for (const unsigned int i : fe_values.dof_indices())
system_rhs(local_dof_indices[i]) += cell_rhs(i);
}
std::map<types::global_dof_index, double> boundary_values;
system_matrix,
solution,
system_rhs);
}
void Step3::solve()
{
}
void Step3::output_results() const
{
std::ofstream output("solution.vtk");
}
{
make_grid();
setup_system();
assemble_system();
solve();
output_results();
}
int main()
{
Step3 laplace_problem;
laplace_problem.run();
return 0;
}
static ::ExceptionBase & ExcNotImplemented()
#define Assert(cond, exc)
void add(const std::vector< size_type > &indices, const std::vector< OtherNumber > &values)
virtual void reinit(const size_type N, const bool omit_zeroing_entries=false)
constexpr const ReferenceCell Quadrilateral
constexpr const ReferenceCell Triangle
Reference cells
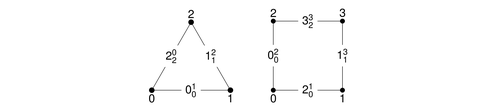
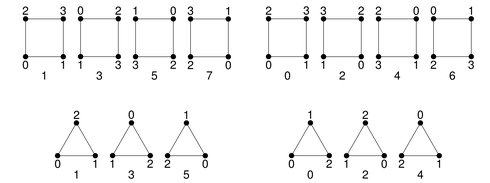
In 2D, we provide triangles and quadrilaterals with the following possible orientations in 3D:
2D: triangle and quadrilateral
Possible orientations of triangles and quadrilaterals in 3D
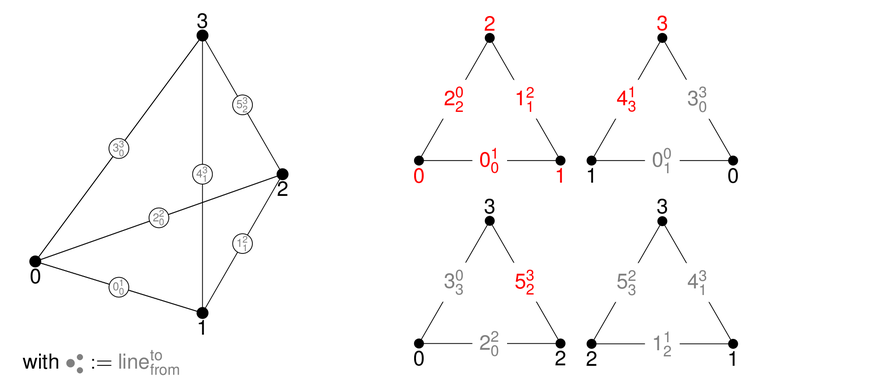
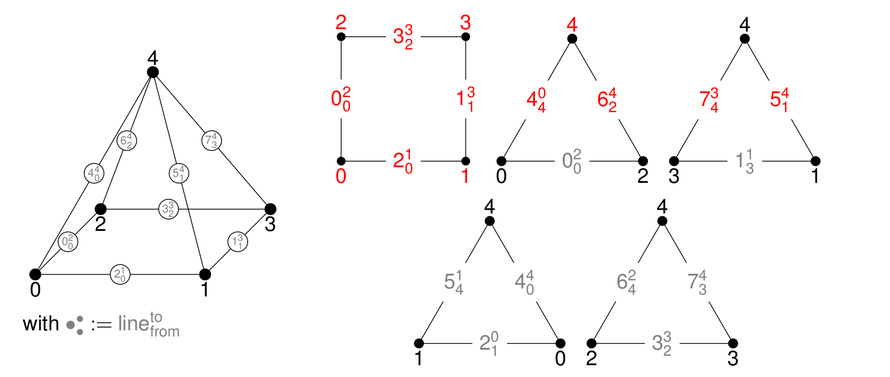
In 3D, tetrahedra, pyramids, wedges, and hexahedra are available:
Each surface of a 3D reference cell consists of 2D reference cells. The documentation of the enumeration of the numbering of their vertices and lines are given in the right columns.
◆ subdivided_hyper_rectangle_with_simplices()
template<
int dim,
int spacedim>
| void GridGenerator::subdivided_hyper_rectangle_with_simplices |
( |
Triangulation< dim, spacedim > & |
tria, |
|
|
const std::vector< unsigned int > & |
repetitions, |
|
|
const Point< dim > & |
p1, |
|
|
const Point< dim > & |
p2, |
|
|
const bool |
colorize = false |
|
) |
| |
Create a coordinate-parallel brick from the two diagonally opposite corner points p1 and p2. The number of vertices in coordinate direction i is given by repetitions[i]+1.
- Note
- This function connects internally 4/8 vertices to quadrilateral/hexahedral cells and subdivides these into 2/5 triangular/tetrahedral cells.
-
Currently, this function only works for
dim==spacedim.
◆ subdivided_hyper_cube_with_simplices()
template<
int dim,
int spacedim>
| void GridGenerator::subdivided_hyper_cube_with_simplices |
( |
Triangulation< dim, spacedim > & |
tria, |
|
|
const unsigned int |
repetitions, |
|
|
const double |
p1 = 0.0, |
|
|
const double |
p2 = 1.0, |
|
|
const bool |
colorize = false |
|
) |
| |
Initialize the given triangulation with a hypercube (square in 2D and cube in 3D) consisting of repetitions cells in each direction. The hypercube volume is the tensor product interval [left,right]^{\text{dim}} in the present number of dimensions, where the limits are given as arguments. They default to zero and unity, then producing the unit hypercube.
- Note
- This function connects internally 4/8 vertices to quadrilateral/hexahedral cells and subdivides these into 2/5 triangular/tetrahedral cells.
◆ read_vtk()
template<
int dim,
int spacedim>
| void GridIn< dim, spacedim >::read_vtk |
( |
std::istream & |
in | ) |
|
Read grid data from a unstructured vtk file. The vtk file may contain the following VTK cell types: VTK_HEXAHEDRON (12), VTK_TETRA (10), VTK_QUAD (9), VTK_TRIANGLE (5), and VTK_LINE (3).
Depending on the template dimension, only some of the above are accepted.
In particular, in three dimensions, this function expects the file to contain
- VTK_HEXAHEDRON/VTK_TETRA cell types
- VTK_QUAD/VTK_TRIANGLE cell types, to specify optional boundary or interior quad faces
- VTK_LINE cell types, to specify optional boundary or interior edges
In two dimensions:
- VTK_QUAD/VTK_TRIANGLE cell types
- VTK_LINE cell types, to specify optional boundary or interior edges
In one dimension
The input file may specify boundary ids, material ids, and manifold ids using the CELL_DATA section of the VTK file format.
This function interprets two types of CELL_DATA contained in the input file: SCALARS MaterialID, used to specify the material_id of the cells, or the boundary_id of the faces and edges, and SCALARS ManifoldID, that can be used to specify the manifold id of any Triangulation object (cell, face, or edge).
The companion GridOut::write_vtk function can be used to write VTK files compatible with this method.
Processing the CELL_TYPES section////////////////////////
Definition at line 128 of file grid_in.cc.
◆ read_msh() [1/2]
template<
int dim,
int spacedim>
| void GridIn< dim, spacedim >::read_msh |
( |
std::istream & |
in | ) |
|
Read grid data from an msh file, either version 1 or version 2 of that file format. The Gmsh formats are documented at http://www.gmsh.info/.
- Note
- The input function of deal.II does not distinguish between newline and other whitespace. Therefore, deal.II will be able to read files in a slightly more general format than Gmsh.
Definition at line 1427 of file grid_in.cc.
◆ read_msh() [2/2]
template<
int dim,
int spacedim>
| void GridIn< dim, spacedim >::read_msh |
( |
const std::string & |
filename | ) |
|
Read grid data using Gmsh API. Any file supported by Gmsh can be passed as argument. The format is deduced from the filename extension.
This function interprets non-named physical ids (gmsh format < 4.0) as material or boundary ids (similarly to what happens with the other read_msh() function). If you want to specify non default manifold or boundary ids, you must group all entities that require a non default boundary or manifold id into named physical groups, where the name is interpreted using the function Patterns::Tools::to_value() applied to a std::map<std::string, int>. The keys can be either MaterialID (if the physical group refers to object of dimension dim), BoundaryID (if the group refers to objects of dimension < dim), or ManifoldID.
From the Gmsh documentation, the formats of the physical tags follows the following conventions:
$PhysicalNames
numPhysicalNames(ASCII int)
dimension(ASCII
int) physicalTag(ASCII
int)
"name"(127 characters
max)
...
$EndPhysicalNames
For example, the following snippet of mesh file
MeshFormat
4.1 0 8
$EndMeshFormat
$PhysicalNames
4
1 1 "ManifoldID:0"
1 2 "BoundaryID: -1, ManifoldID: 1"
2 3 "ManifoldID: 1"
2 4 "MaterialID: 2, ManifoldID: 1"
$EndPhysicalNames
$Entities
...
refers to a two dimensional grid where:
- a portion of the boundary of dimension 1 has physical tag 1, and manifold id 0
- some internal faces (lines of dimension 1) have manifold id 1
- some elements have manifold id 1 (and material id equal to the default value, i.e., zero)
- some elements have manifold id 1 and material id equal to 2
If the physical groups are not named, then the behaviour is the same as the other read_msh() function, i.e., the physical tag itself is interpreted as a boundary or material id.
Definition at line 2124 of file grid_in.cc.