This program was contributed by Shamil Magomedov <magomedov.shamil.m@gmail.com>.
It comes without any warranty or support by its authors or the authors of deal.II.
This program is part of the deal.II code gallery and consists of the following files (click to inspect):
Pictures from this code gallery program
Annotated version of README.md
Traveling-wave solutions of a qualitative model for combustion waves
This program demonstrates the use of adaptive finite elements to compute traveling-wave solutions of partial differential equations. One of the challenges in solving this type of problem is the presence of an unknown wave speed. Below we show how to overcome this.
Building, compiling and running
To run the program, enter the following commands from the current directory:
mkdir build && cd build
cmake ..
make
./main ../ParametersList.prm
Problem
To illustrate the algorithm for the computation of traveling-wave profiles, we consider a combustion model described in [1]. In a moving reference frame of a steadily propagating wave we have the following nondimensionalized system:
\begin{align*}
- c u_{\xi} + (1 + \epsilon u) u_{\xi} &= -\dfrac{\epsilon}{2} T_{\xi} + \dfrac{4 \delta}{3 \epsilon} \Pr u_{\xi \xi}, \\
- c(T_{\xi} - u_{\xi}) &= q \omega + \delta T_{\xi \xi}, \\
- c \lambda_{\xi} &= \omega + \dfrac{\delta}{\text{Le}} \lambda_{\xi \xi}.
\end{align*}
Here, u is the pressure, T is the temperature, \lambda is the reaction-progress variable, varying from 0 in the fresh mixture to 1 in the burnt products, c > 0 is the unknown wave speed, \Pr and \mathrm{Le} are the Prandtl and Lewis numbers, q is the energy of heat release. The model parameters \epsilon and \delta determine the strength of nonlinearity and dissipative effects. The reaction rate \omega is taken as
\begin{align*}
\omega = k (1 - \lambda) \exp(-\theta / T) \, \mathrm{H}(T - T_{\mathrm{ign}}),
\end{align*}
with activation energy \theta, ignition temperature T_{\mathrm{ign}}, constant of the reaction rate k, and the Heaviside step function \mathrm{H}.
The boundary conditions at \xi = -\infty are
\begin{align*}
u_{\xi} = 0, \ T = T_l, \ \lambda = 1,
\end{align*}
and at \xi = +\infty are
\begin{align*}
u = u_r, \ \lambda = 0.
\end{align*}
The right boundary condition for temperature is T = T_r for detonation waves (supersonic regime, i.e. c > 1) and T_{\xi} = 0 for deflagration waves (subsonic regime, i.e. c < 1).
Because of translational invariance, we need to impose another constraint on the system to fix a particular solution. So we choose the following centering condition: T(0) = T_{\mathrm{ign}}.
Numerical algorithm
Newton–Raphson iteration scheme
The nonlinear boundary value problem is solved numerically on a finite interval I = [l, r] \left(|l|, |r| \gg 1 \right), using a Newton–Raphson iteration scheme, similar to one, described in deal.II tutorial step-15. The main difference from step-15 is that we have an additional scalar unknown, the front velocity c. So the algorithm has to be modified to take this feature into account.
Rewriting the system in a vector form
\begin{align*}
\mathbf{F}(u, T, \lambda, c) =
\left(\begin{array}{c}
\dfrac{4 \delta}{3 \epsilon} \Pr u_{\xi \xi} - u_{\xi} (1 - c + \epsilon u) -\dfrac{\epsilon}{2} T_{\xi} \\[7pt]
\delta T_{\xi \xi} + c(T_{\xi} - u_{\xi}) + q \omega \\[5pt]
\dfrac{\delta}{\text{Le}} \lambda_{\xi \xi} + c \lambda_{\xi} + \omega
\end{array} \right)
= 0,
\end{align*}
we define a Newton–Raphson iteration as
\begin{align*}
\mathbf{F'}(\mathbf{x}^k, \mathbf{dx}^k) = - \mathbf{F}(\mathbf{x}^k),
\end{align*}
\begin{align*}
\mathbf{x}^{n+1} = \mathbf{x}^{n} + \alpha^k \mathbf{dx}^k,
\end{align*}
where k is the step number, \mathbf{x}^k = (u^k, T^k, \lambda^k, c^k)^{\top} is a vector argument, \mathbf{dx}^k = (du^k, dT^k, d\lambda^k, dc^k)^{\top} is an increment, \alpha^k is some damping parameter for managing the global convergence behavior and \mathbf{F'}(\mathbf{x}^k, \mathbf{dx}^k) is the directional derivative, defined as
\begin{align*}
\mathbf{F'}(\mathbf{x}, \mathbf{dx}) = \dfrac{\mathrm{d}}{\mathrm{d} \varepsilon} \Big|_{\varepsilon=0} \mathbf{F}(\mathbf{x} &+ \varepsilon \mathbf{dx}).
\end{align*}
The system to be solved at every iteration step to obtain the increment \mathbf{dx}^k = (du^k, dT^k, d\lambda^k, dc^k)^{\top} can be represented in matrix vector notation as follows
\begin{align*}
\begin{pmatrix}
\dfrac{4 \delta}{3 \epsilon} \Pr \partial_{\xi \xi} - (1 - c + \epsilon u)\partial_{\xi} - \epsilon u_{\xi} & -\dfrac{\epsilon}{2} \partial_{\xi} & 0 & u_{\xi} \\[9pt]
-c \partial_{\xi} & \delta \partial_{\xi \xi} + c \partial_{\xi} + q \kappa_1 & q \kappa_2 & T_{\xi} - u_{\xi} \\[9pt]
0 & \kappa_1 & \dfrac{\delta}{\text{Le}} \partial_{\xi \xi} + c \partial_{\xi} + \kappa_2 & \lambda_{\xi}
\end{pmatrix}
\begin{pmatrix}
du \\[9pt]
dT \\[9pt]
d\lambda \\[9pt]
dc
\end{pmatrix} = -\begin{pmatrix}
f_1 \\[9pt]
f_2 \\[9pt]
f_3
\end{pmatrix},
\end{align*}
where
\begin{align*}
\kappa_1 &= k (1 - \lambda) \exp(-\theta / T) \left[ \dfrac{\theta }{T^2} \, \text{H}(T - T_{\text{ign}}) + \delta(T - T_{\text{ign}}) \right], \\
\kappa_2 &= - k \exp(-\theta / T) \, \text{H}(T - T_{\text{ign}}),
\end{align*}
in which \delta(\cdot) is a Dirac delta function, and f_i \, (i=1,2,3) are the components of the vector function \mathbf{F}(u, T, \lambda, c). The term \delta(T - T_{\text{ign}}) can be rewritten as
\begin{align*}
\delta(T - T_{\text{ign}}) = \frac{\delta(\xi)}{|T'(0)|}.
\end{align*}
We choose the initial guess \mathbf{x}^0 to include the appropriate boundary values, therefore the update \mathbf{dx}^k uses homogeneous Dirichlet or Neumann boundary conditions.
Weak formulation
We multiply both sides of the equation for \mathbf{dx}^k with vector valued test function \mathbf{v} = (v_1, v_2, v_3)^{\top} and integrate over the domain \Omega to obtain a scalar equation
\begin{align*}
J(\mathbf{dx}, \mathbf{v}) = -b(\mathbf{v}).
\end{align*}
\begin{align*}
J(\mathbf{dx}, &\mathbf{v}) = \int \limits_{\Omega} \mathbf{v} \cdot \mathbf{F'}(\mathbf{x}, \mathbf{dx}) \, d\xi = \\
= &\dfrac{4 \delta}{3 \epsilon} \Pr (-\partial_{\xi} v_1, \partial_{\xi} du) + (v_1, - (1 - c + \epsilon u)\partial_{\xi} du - \epsilon u_{\xi} du -\dfrac{\epsilon}{2} \partial_{\xi} dT + u_{\xi} dc) + \\
&+ \delta (-\partial_{\xi} v_2, \partial_{\xi} dT) + (v_2, -c \, \partial_{\xi} du + c \, \partial_{\xi} dT + q \kappa_1 dT + q \kappa_2 d\lambda + T_{\xi} dc - u_{\xi} dc) + \\
&+ \dfrac{\delta}{\text{Le}} (-\partial_{\xi} v_3, \partial_{\xi} d\lambda) + (v_3, \kappa_1 dT + c \partial_{\xi} d\lambda + \kappa_2 d\lambda + \lambda_{\xi} dc).
\end{align*}
\begin{align*}
b(\mathbf{v}) = \int \limits_{\Omega} &\mathbf{v} \cdot \mathbf{F}(\mathbf{x}) \, d\xi = \\
= &\dfrac{4 \delta}{3 \epsilon} \Pr (-\partial_{\xi} v_1, u_{\xi}) + (v_1, - u_{\xi} (1 - c + \epsilon u) -\dfrac{\epsilon}{2} T_{\xi}) + \\
&+ \delta (-\partial_{\xi} v_2, T_{\xi}) + (v_2, c(T_{\xi} - u_{\xi}) + q \omega) + \\
&+ \dfrac{\delta}{\text{Le}} (-\partial_{\xi} v_3, \lambda_{\xi}) + (v_3, c \lambda_{\xi} + \omega).
\end{align*}
In the above expressions second derivatives disappear due to integration by parts with homogeneous Dirichlet and Neumann boundary conditions. The solution is sought as an expansion
\begin{align*}
\begin{pmatrix}
du \\[9pt]
dT \\[9pt]
d\lambda \\[9pt]
dc
\end{pmatrix} =
\sum \limits_{i = 1}^{3N}
U_{i}\begin{pmatrix}
\phi_i^1 \\[9pt]
\phi_i^2 \\[9pt]
\phi_i^3 \\[9pt]
0
\end{pmatrix} +
U_{3N + 1}\begin{pmatrix}
0 \\[9pt]
0 \\[9pt]
0 \\[9pt]
1
\end{pmatrix} \quad \in \quad V_p \times V_p \times V_p \times \mathbb{R}.
\end{align*}
where V_p is a finite element space of continuous, piecewise polynomials of degree p. The set of vector functions (\phi_i^1, \phi_i^2, \phi_i^3)^{\top} \in V_p^3 form the basis of the corresponding space. We then choose test functions \mathbf{v} to be the same as the basis functions, and obtain the linear system J U = b. Elements of the matrix and the right-hand side are computed according to following formulas:
\begin{align*}
J_{ij} = &\dfrac{4 \delta}{3 \epsilon} \Pr (-\partial_{\xi} \phi_i^1, \partial_{\xi} \phi_j^1) + (\phi_i^1, - (1 - c + \epsilon u)\partial_{\xi} \phi_j^1 - \epsilon u_{\xi} \phi_j^1 -\dfrac{\epsilon}{2} \partial_{\xi} \phi_j^2) + \\
&+ \delta (-\partial_{\xi} \phi_i^2, \partial_{\xi} \phi_j^2) + (\phi_i^2, -c \, \partial_{\xi} \phi_j^1 + c \, \partial_{\xi} \phi_j^2 + q \kappa_1 \phi_j^2 + q \kappa_2 \phi_j^3) + \\
&+ \dfrac{\delta}{\text{Le}} (-\partial_{\xi} \phi_i^3, \partial_{\xi} \phi_j^3) + (\phi_i^3, \kappa_1 \phi_j^2 + c \partial_{\xi} \phi_j^3 + \kappa_2 \phi_j^3),
\end{align*}
\begin{align*}
J_{i, 3N + 1} = (\phi_i^1, u_{\xi}) + (\phi_i^2, T_{\xi} - u_{\xi}) + (\phi_i^3, \lambda_{\xi}),
\end{align*}
\begin{align*}
b_{i} = &\dfrac{4 \delta}{3 \epsilon} \Pr (-\partial_{\xi} \phi_i^1, u_{\xi}) + (\phi_i^1, - u_{\xi} (1 - c + \epsilon u) -\dfrac{\epsilon}{2} T_{\xi}) + \\
&+ \delta (-\partial_{\xi} \phi_i^2, T_{\xi}) + (\phi_i^2, c(T_{\xi} - u_{\xi}) + q \omega) + \\
&+ \dfrac{\delta}{\text{Le}} (-\partial_{\xi} \phi_i^3, \lambda_{\xi}) + (\phi_i^3, c \lambda_{\xi} + \omega),
\end{align*}
for i, j < 3N + 1.
In order for the system to have a unique solution, we need to supplement it with one more equation, corresponding to the constraint T(0) = T_{\text{ign}}. The initial approximation to the solution is set so as to satisfy this condition, so we just need the computed increment function dT to be zero at the specified point. Thus, we add a row of zeros with a value of 1 in the position corresponding to dT(0) to the matrix J and set b_{3N + 1} = 0.
The resulting sparsity pattern structure has the form shown in the figure below. 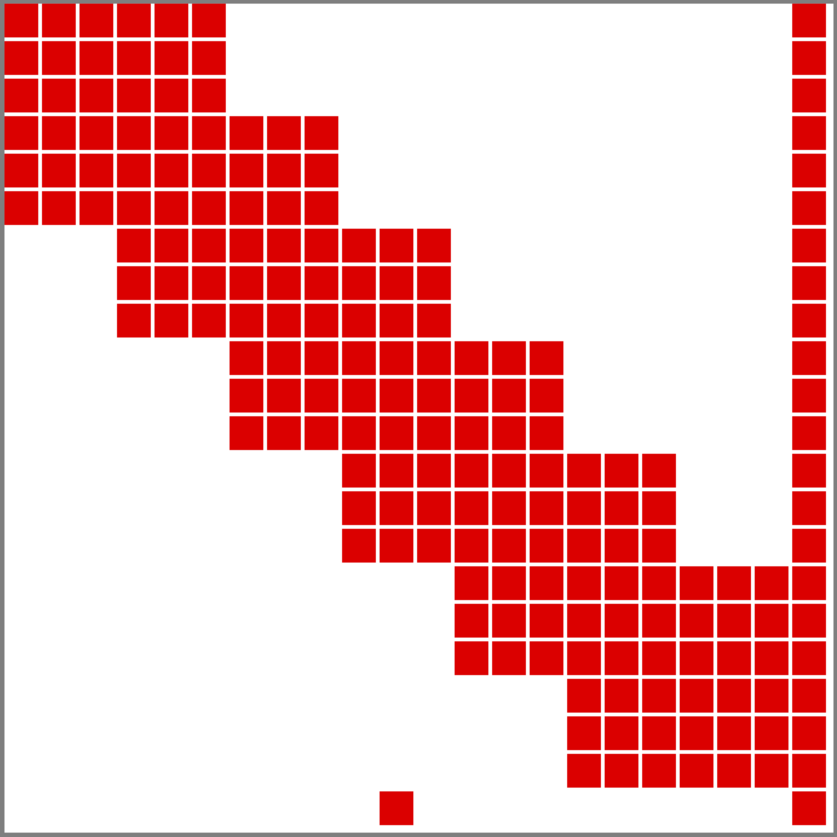
The integration of the terms with \kappa_1 need special attention because of the Dirac delta function. If U_n and U_m are the degrees of freedom, associated with the vertex \xi = 0 (i.e., \phi_n^2(0) = 1 and \phi_m^3(0) = 1), we get
\begin{align*}
(\phi_n^2, q (k (1 - \lambda) \exp(-\theta / T) \delta(T - T_{\text{ign}})) \phi_n^2) = \dfrac{q k (1 - \lambda(0)) \exp(-\theta / T(0))}{|T'(0)|}
\end{align*}
and
\begin{align*}
(\phi_m^3, (k (1 - \lambda) \exp(-\theta / T) \delta(T - T_{\text{ign}})) \phi_n^2) = \dfrac{k (1 - \lambda(0)) \exp(-\theta / T(0))}{|T'(0)|}.
\end{align*}
Initial guess
The initial guess for detonation wave is obtained from the following problem
\begin{align*}
u_{\xi} (- c + 1 + \epsilon u) &= -\dfrac{\epsilon}{2} T_{\xi} , \\
- c(T_{\xi} - u_{\xi}) &= q \omega, \\
- c \lambda_{\xi} &= \omega,
\end{align*}
which is the limiting case of the system at \delta = 0. The problem reduces to the nonlinear initial value problem
\begin{align*}
\lambda_{\xi} = -\dfrac{k}{c} (1 - \lambda) \exp \left( \dfrac{-\theta}{T(\lambda)} \right),
\end{align*}
with initial condition \lambda(0) = 0; see [1] for details.
For the deflagration case, the initial guess is taken piecewise constant for u and T, and
\begin{align*}
\lambda(\xi) =
\begin{cases}
-\exp \left(\xi (1 - c) \Big/ \left(\dfrac{4 \delta}{3 \epsilon} \Pr \right) \right) + 1 \quad &\mathrm{for}\ \xi \in [l, 0], \\
0 \quad &\mathrm{for}\ \xi \in (0, r]
\end{cases}
\end{align*}
for the reaction-progress variable. The value in the interval (0, 1) is chosen as the initial guess for the front velocity c.
Boundary conditions
In the numerical solution, the boundary conditions described in the beginning are imposed at the ends of the interval I. In addition, a homogeneous Neumann condition is applied to the function d\lambda at the left boundary.
Program
Parameters
The calculation parameters are set in the ParametersList.prm file. To reproduce the results obtained below, you can run the program with the parameter files ParametersListDeflagrationSlow.prm, ParametersListDeflagrationFast.prm and ParametersListDetonation.prm.
Class TravelingWaveSolver
TravelingWaveSolver is the main class for computation of the traveling-wave profiles.
The implementation of Newton's method is based on that described in step-77 and relies on SUNDIALS' KINSOL package. Because of the additional unknown, the front velocity, we expand the Jacobi matrix by one column and one row (jacobian_matrix_extended), and add one more element to the solution vector (current_solution_extended). After completing the Newton iterations, we split the resulting extended solution vector current_solution_extended into two parts: the solution vector current_solution, corresponding to (u, T, \lambda), and the front velocity current_wave_speed. After that the adaptive mesh refinement is performed using the current_solution vector, which is very important for resolving a narrow transition layer with a large solution gradient in the vicinity of zero. The KellyErrorEstimator is used as a refinement indicator.
Function calculate_profile()
The full calculation cycle is done in the calculate_profile function. First, we construct an initial guess to the solution depending on the selected wave type and store the result as an object of type SolutionStruct. This object, along with the problem parameters, is then passed to the constructor of the TravelingWaveSolver class to calculate the traveling wave.
Decreasing the dissipation parameter \delta leads to the appearance of large gradients in solutions in the neighborhood of zero. As a consequence, Newton's method becomes more sensitive to the initial data and ceases to converge. To solve this problem, the calculate_profile function implements the method of continuation by the \delta parameter (for an example, see step-57). The solution and the refined triangulation are saved after each step of the method using the get_solution and get_triangulation functions and then passed to the next step.
Error estimation
Integration of the governing equations over the real line gives the following relations:
\begin{align*}
u_l (1 - c) + \frac{\epsilon}{2} u_l^2 + \frac{\epsilon}{2} T_l &= u_r (1 - c) + \frac{\epsilon}{2} u_r^2 + \frac{\epsilon}{2} T_r , \\
T_l - u_l &= T_r - u_r + q.
\end{align*}
These relations let us express any two parameters of c, T_l, T_r, u_l, u_r in terms of the remaining three. Thus, we can write
\begin{align*}
u_l &= (T_l - T_r) + u_r - q, \\
c &= 1 + \epsilon \left( u_r - \dfrac{(q - (T_l - T_r))^2 + (T_l - T_r)}{2 (q - (T_l - T_r))} \right).
\end{align*}
This means that since we choose the three parameters T_l, T_r, u_r for the detonation case ourselves, the above formulas give us the exact values of c and u_l. These can be used to obtain the value of the error in the calculated c and u_l.
For the deflagration case, however, we can only choose two parameters, T_l and u_r. The remaining three are determined during the solution, so the formulas can only give us an error estimate.
Initial guess
To get the initial condition for detonation, we have to solve the nonlinear initial value problem for \lambda we mentioned earlier. This is done in the LimitSolution class. Numerical integration is performed using the odeint library of the Boost with its interface in IntegrateSystem.h.
Results
To visualize the computed profiles, one can use gnuplot typing
plot for [i=2:4] "solution_filename" using 1:i w p title word("u T lambda", i-1)
or execute the python script plot.py
python plot.py "solution_filename"
Slow deflagration for delta = 0.01
The calculated wave speed is c = 0.0909. 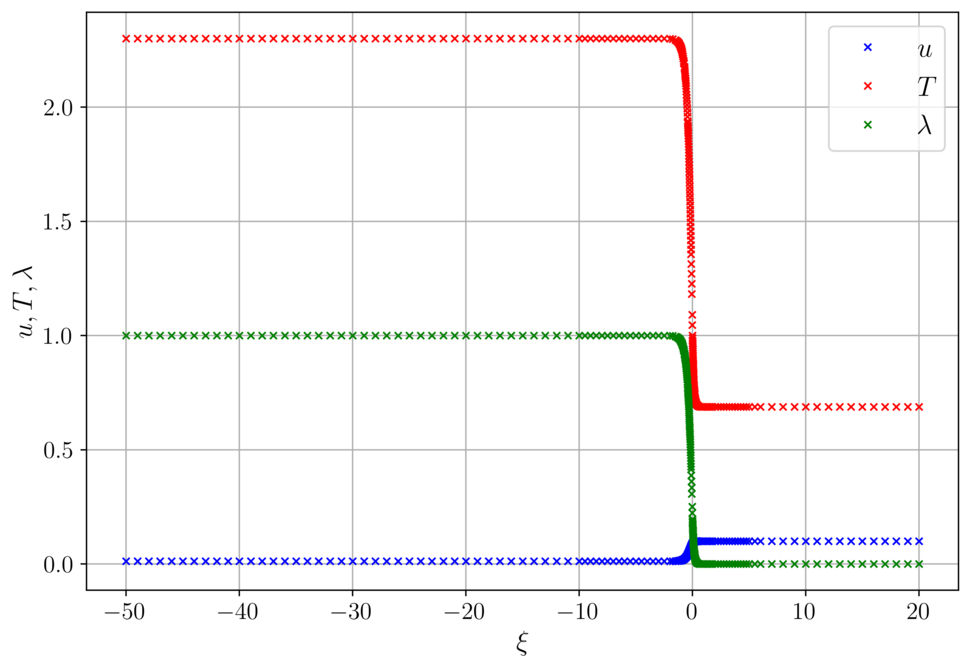
Fast deflagration for delta = 0.01
The calculated wave speed is c = 0.8252. 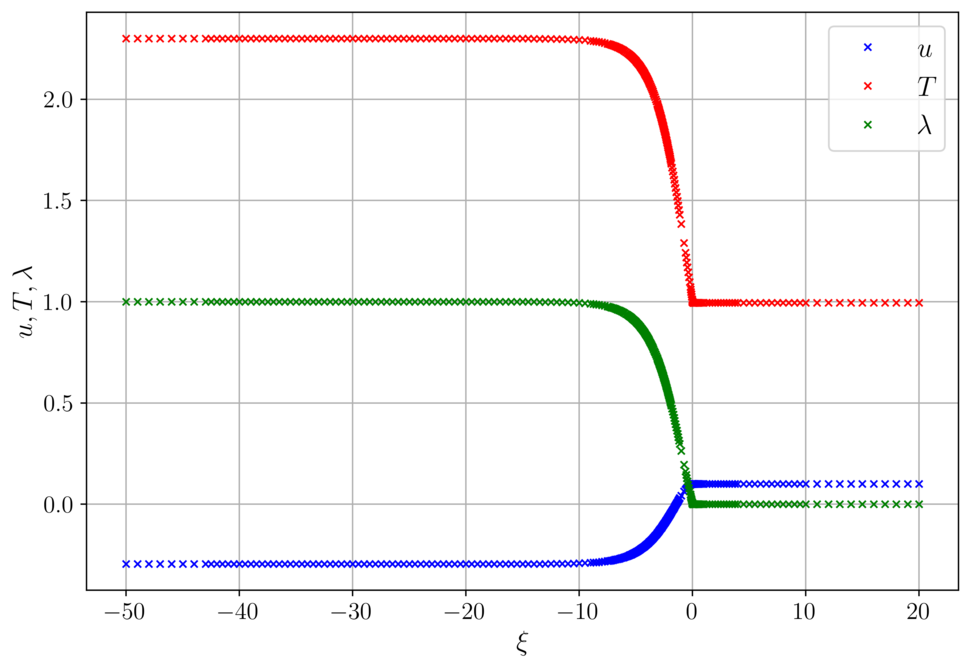
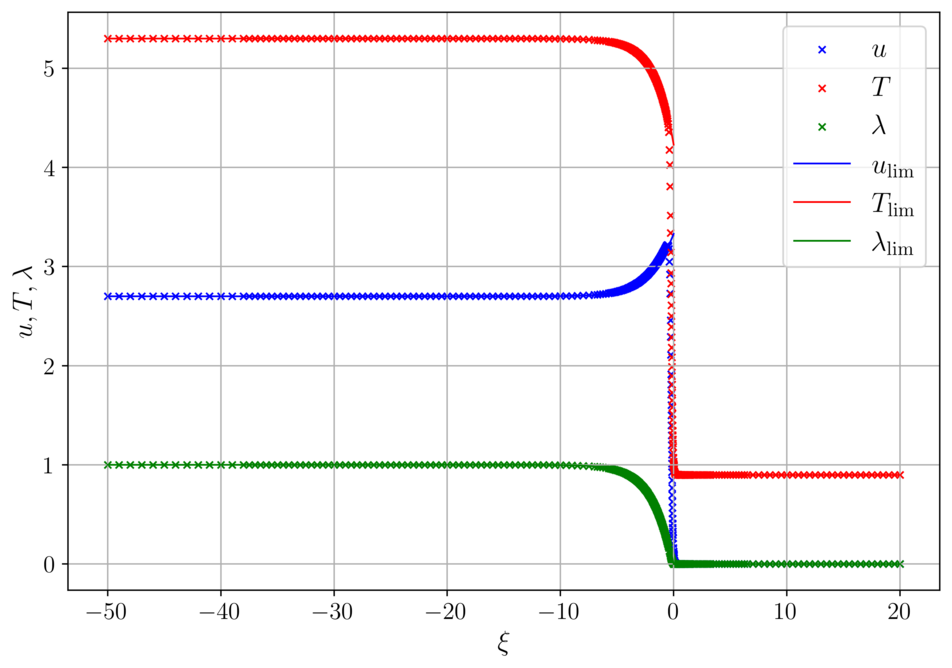
Detonation for delta = 0.01 and delta = 0.001
The calculated wave speed in both cases is the same c = 1.216481, as expected. Solid lines represent the detonation profile for the ideal case, when \delta=0.
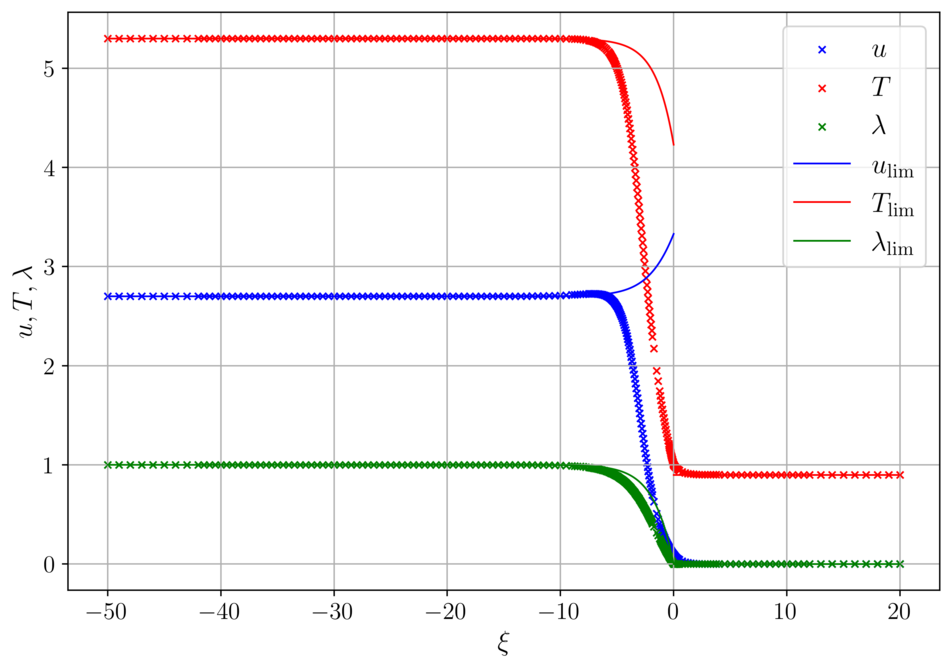
Acknowledgments
I would like to thank my friend Oleg Rogozin for introducing me to the deal.II library and the world of finite elements.
References
- Goldin A.Y., Magomedov S.M., Faria L.M., Kasimov A.R. Study of a qualitative model for combustion waves: Flames, detonations, and deflagration-to-detonation transition. Computers & Fluids 2024; 273:106213.
Annotated version of AuxiliaryFunctions.h
#ifndef AUXILIARY_FUNCTIONS
#define AUXILIARY_FUNCTIONS
#include <vector>
#include <cmath>
#include <fstream>
#include <string>
#include <cstring>
Comparison of numbers with a given tolerance.
template <typename T>
bool isapprox(const T &a, const T &b, const double tol = 1e-10)
{
}
::VectorizedArray< Number, width > abs(const ::VectorizedArray< Number, width > &)
Fill the std::vector with the values from the range [interval_begin, interval_end].
template <typename T>
void linspace(T interval_begin, T interval_end, std::vector<T> &arr)
{
const size_t SIZE = arr.size();
const T step = (interval_end - interval_begin) / static_cast<T>(SIZE - 1);
for (size_t i = 0; i < SIZE; ++i)
{
arr[i] = interval_begin + i * step;
}
}
Check the file existence.
inline bool file_exists(const std::string &filename)
{
std::ifstream f(filename.c_str());
return f.good();
}
#endif
Annotated version of IntegrateSystem.h
#ifndef INTEGRATE_SYSTEM
#define INTEGRATE_SYSTEM
#include <
boost/numeric/odeint.hpp>
#include <iostream>
#include <fstream>
#include <string>
template <typename state_T, typename time_T>
void SaveSolutionIntoFile(const std::vector<state_T>& x_vec, const std::vector<time_T>& t_vec, std::string filename="output_ode_sol.txt")
{
if (!x_vec.empty() && !t_vec.empty())
{
std::ofstream output(filename);
output << std::setprecision(16);
size_t dim = x_vec[0].size();
for (size_t i = 0; i < t_vec.size(); ++i)
{
output << std::fixed << t_vec[i];
for (size_t j = 0; j < dim; ++j)
{
output << std::scientific << " " << x_vec[i][j];
}
output << "\n";
}
output.close();
}
else
{
std::cout << "Solution is not saved into file.\n";
}
}
type of RK integrator
enum class Integrator_Type
{
dopri5,
cash_karp54,
fehlberg78
};
Observer
template <typename state_type>
class Push_back_state_time
{
public:
std::vector<state_type>& m_states;
std::vector<double>& m_times;
Push_back_state_time(std::vector<state_type>& states, std::vector<double>& times)
: m_states(states), m_times(times)
{}
void operator() (const state_type& x, double t)
{
m_states.push_back(x);
m_times.push_back(t);
}
};
Integrate system at specified points.
template <typename ODE_obj_T, typename state_type, typename Iterable_type>
void IntegrateSystemAtTimePoints(
std::vector<state_type>& x_vec, std::vector<double>& t_vec, const Iterable_type& iterable_time_span,
const ODE_obj_T& ode_system_obj,
state_type& x, const double dt,
Integrator_Type integrator_type=Integrator_Type::dopri5, bool save_to_file_flag=false,
const double abs_er_tol=1.0e-15, const double rel_er_tol=1.0e-15
)
{
using namespace boost::numeric::odeint;
if (integrator_type == Integrator_Type::dopri5)
{
typedef runge_kutta_dopri5< state_type > error_stepper_type;
integrate_times( make_controlled< error_stepper_type >(abs_er_tol, rel_er_tol),
ode_system_obj, x, iterable_time_span.begin(), iterable_time_span.end(), dt, Push_back_state_time< state_type >(x_vec, t_vec) );
}
else if (integrator_type == Integrator_Type::cash_karp54)
{
typedef runge_kutta_cash_karp54< state_type > error_stepper_type;
integrate_times( make_controlled< error_stepper_type >(abs_er_tol, rel_er_tol),
ode_system_obj, x, iterable_time_span.begin(), iterable_time_span.end(), dt, Push_back_state_time< state_type >(x_vec, t_vec) );
}
else
{
typedef runge_kutta_fehlberg78< state_type > error_stepper_type;
integrate_times( make_controlled< error_stepper_type >(abs_er_tol, rel_er_tol),
ode_system_obj, x, iterable_time_span.begin(), iterable_time_span.end(), dt, Push_back_state_time< state_type >(x_vec, t_vec) );
}
if (save_to_file_flag)
{
SaveSolutionIntoFile(x_vec, t_vec);
}
}
#endif
Annotated version of LimitSolution.cc
#include "LimitSolution.h"
namespace TravelingWave
{
LimitSolution::LimitSolution(const Parameters ¶meters, const double ilambda_0, const double iu_0, const double iT_0, const double iroot_sign)
: params(parameters)
, problem(params.problem)
, wave_speed(problem.wave_speed_init)
, lambda_0(ilambda_0)
, u_0(iu_0)
, T_0(iT_0)
, root_sign(iroot_sign)
{
calculate_constants_A_B();
}
double LimitSolution::omega_func(const double lambda, const double T) const
{
return problem.k * (1. - lambda) *
std::exp(-problem.theta / T);
}
void LimitSolution::operator() (const state_type &x , state_type &dxdt , const double )
{
dxdt[0] = -1. / wave_speed * omega_func(x[0], T_func(x[0]));
}
double LimitSolution::u_func(const double lambda) const
{
double coef = 2 * (wave_speed - 1) / problem.epsilon - 1;
return (coef + root_sign *
std::sqrt(coef * coef - 4 * (problem.q * lambda + B - 2 * A / problem.epsilon))) / 2;
}
double LimitSolution::T_func(const double lambda) const
{
return u_func(lambda) + problem.q *
lambda + B;
}
void LimitSolution::calculate_constants_A_B()
{
B = T_0 - u_0;
A = u_0 * (1 - wave_speed) + problem.epsilon * (u_0 * u_0 + T_0) / 2;
}
void LimitSolution::set_wave_speed(double iwave_speed)
{
wave_speed = iwave_speed;
calculate_constants_A_B();
}
void LimitSolution::calculate_u_T_omega()
{
if (!t_vec.empty() && !lambda_vec.empty())
{
u_vec.resize(lambda_vec.size());
T_vec.resize(lambda_vec.size());
omega_vec.resize(lambda_vec.size());
for (unsigned int i = 0; i < lambda_vec.size(); ++i)
{
u_vec[i].resize(1);
T_vec[i].resize(1);
omega_vec[i].resize(1);
u_vec[i][0] = u_func(lambda_vec[i][0]);
T_vec[i][0] = T_func(lambda_vec[i][0]);
omega_vec[i][0] = omega_func(lambda_vec[i][0], T_vec[i][0]);
}
}
else
{
std::cout << "t_vec or lambda_vec vector is empty!" << std::endl;
}
}
}
::VectorizedArray< Number, width > exp(const ::VectorizedArray< Number, width > &)
::VectorizedArray< Number, width > sqrt(const ::VectorizedArray< Number, width > &)
Annotated version of LimitSolution.h
#ifndef LIMIT_SOLUTION
#define LIMIT_SOLUTION
#include "Parameters.h"
#include <iostream>
#include <vector>
namespace TravelingWave
{
typedef std::vector< double > state_type;
class LimitSolution
{
public:
LimitSolution(const Parameters ¶meters, const double ilambda_0, const double iu_0, const double iT_0, const double root_sign = 1.);
void operator() (const state_type &x , state_type &dxdt , const double );
void calculate_u_T_omega();
void set_wave_speed(double iwave_speed);
std::vector<double> t_vec;
std::vector<state_type> omega_vec;
std::vector<state_type> lambda_vec;
std::vector<state_type> u_vec;
std::vector<state_type> T_vec;
private:
double omega_func(const double lambda, const double T) const;
double u_func(const double lambda) const;
double T_func(const double lambda) const;
void calculate_constants_A_B();
const Parameters ¶ms;
const Problem &problem;
double wave_speed;
const double lambda_0, u_0, T_0;
double A, B;
const double root_sign;
};
}
#endif
Annotated version of LinearInterpolator.h
#ifndef LINEAR_INTERPOLATOR
#define LINEAR_INTERPOLATOR
#include <cmath>
#include <algorithm>
#include <vector>
Linear interpolation class
template <typename Number_Type>
class LinearInterpolator
{
public:
LinearInterpolator(const std::vector<Number_Type> &ix_points, const std::vector<Number_Type> &iy_points);
Number_Type value(const Number_Type x) const;
private:
const std::vector<Number_Type> x_points;
const std::vector<Number_Type> y_points;
};
template <typename Number_Type>
LinearInterpolator<Number_Type>::LinearInterpolator(const std::vector<Number_Type> &ix_points, const std::vector<Number_Type> &iy_points)
: x_points(ix_points)
, y_points(iy_points)
{}
template <typename Number_Type>
Number_Type LinearInterpolator<Number_Type>::value(const Number_Type x) const
{
Number_Type res = 0.;
auto lower = std::lower_bound(x_points.begin(), x_points.end(), x);
unsigned int right_index = 0;
unsigned int left_index = 0;
if (lower == x_points.begin())
{
res = y_points[0];
}
else if (lower == x_points.end())
{
res = y_points[x_points.size()-1];
}
else
{
right_index = lower - x_points.begin();
left_index = right_index - 1;
Number_Type y_2 = y_points[right_index];
Number_Type y_1 = y_points[left_index];
Number_Type x_2 = x_points[right_index];
Number_Type x_1 = x_points[left_index];
res = (y_2 - y_1) / (x_2 - x_1) * (x - x_1) + y_1;
}
return res;
}
#endif
Annotated version of Parameters.cc
#include "Parameters.h"
namespace TravelingWave
{
Problem::Problem()
{
add_parameter("delta", delta = 0.01);
add_parameter("epsilon", epsilon = 0.1);
add_parameter("Prandtl number", Pr = 0.75);
add_parameter("Lewis number", Le = 1.0);
add_parameter("Constant of reaction rate", k = 1.0);
add_parameter("Activation energy", theta = 1.65);
add_parameter("Heat release", q = 1.7);
add_parameter("Ignition Temperature", T_ign = 1.0);
add_parameter("Type of the wave (deflagration / detonation)", wave_type = 1);
add_parameter("Type of boundary condition for the temperature at the right boundary", T_r_bc_type = 1);
add_parameter("T_left", T_left = 5.3);
add_parameter("T_right", T_right = 0.9);
add_parameter("u_left", u_left = -0.2);
add_parameter("u_right", u_right = 0.);
add_parameter("Initial guess for the wave speed", wave_speed_init = 1.2);
}
FiniteElements::FiniteElements()
{
add_parameter("Polynomial degree", poly_degree = 1);
add_parameter("Number of quadrature points", quadrature_points_number = 3);
}
Mesh::Mesh()
{
add_parameter("Interval left boundary", interval_left = -50.0);
add_parameter("Interval right boundary", interval_right = 20.0);
add_parameter<unsigned int>("Refinements number", refinements_number = 10);
add_parameter("Adaptive mesh refinement", adaptive = 1);
}
Solver::Solver()
{
add_parameter("Tolerance", tol = 1e-10);
}
}
Annotated version of Parameters.h
#ifndef PARAMETERS
#define PARAMETERS
#include <deal.II/base/parameter_acceptor.h>
namespace TravelingWave
{
{
Problem();
double delta, epsilon;
double Pr, Le;
double k, theta, q;
double T_ign;
int wave_type;
int T_r_bc_type;
double T_left, T_right;
double u_left, u_right;
double wave_speed_init;
};
{
FiniteElements();
unsigned int poly_degree;
unsigned int quadrature_points_number;
};
{
Mesh();
double interval_left;
double interval_right;
unsigned int refinements_number;
int adaptive;
};
{
Solver();
double tol;
};
struct Parameters
{
Problem problem;
FiniteElements fe;
Mesh mesh;
Solver solver;
};
}
#endif
Annotated version of Solution.cc
#include "Solution.h"
namespace TravelingWave
{
SolutionStruct::SolutionStruct() {}
SolutionStruct::SolutionStruct(const std::vector<double> &ix, const std::vector<double> &iu,
const std::vector<double> &iT, const std::vector<double> &ilambda, double iwave_speed)
: x(ix)
, u(iu)
, T(iT)
, lambda(ilambda)
, wave_speed(iwave_speed)
{}
SolutionStruct::SolutionStruct(const
std::vector<double> &ix, const
std::vector<double> &iu,
const
std::vector<double> &iT, const
std::vector<double> &ilambda)
: SolutionStruct(ix, iu, iT, ilambda, 0.)
{}
void SolutionStruct::reinit(const unsigned
int number_of_elements)
{
wave_speed = 0.;
x.clear();
u.clear();
T.clear();
lambda.clear();
x.resize(number_of_elements);
u.resize(number_of_elements);
T.resize(number_of_elements);
lambda.resize(number_of_elements);
}
void SolutionStruct::save_to_file(std::string filename = "sol") const
{
const std::string file_for_solution = filename + ".txt";
std::ofstream output(file_for_solution);
output << std::scientific << std::setprecision(16);
for (unsigned int i = 0; i < x.size(); ++i)
{
output << std::fixed << x[i];
output << std::scientific <<
" " << u[i] <<
" " << T[i] <<
" " <<
lambda[i] <<
"\n";
}
output.close();
std::ofstream file_for_wave_speed_output("wave_speed-" + file_for_solution);
file_for_wave_speed_output << std::scientific << std::setprecision(16);
file_for_wave_speed_output << wave_speed << std::endl;
file_for_wave_speed_output.close();
}
Interpolant::Interpolant(const std::vector<double> &ix_points, const std::vector<double> &iy_points)
: interpolant(ix_points, iy_points)
{}
double Interpolant::
value(const
Point<1> &p, const unsigned
int component) const
{
double x = p[0];
double res = interpolant.value(x);
return res;
}
template <typename InterpolantType>
SolutionVectorFunction<InterpolantType>::SolutionVectorFunction(InterpolantType iu_interpolant, InterpolantType iT_interpolant, InterpolantType ilambda_interpolant)
, u_interpolant(iu_interpolant)
, T_interpolant(iT_interpolant)
, lambda_interpolant(ilambda_interpolant)
{}
template <typename InterpolantType>
double SolutionVectorFunction<InterpolantType>::
value(const
Point<1> &p, const unsigned
int component) const
{
double res = 0.;
if (component == 0) { res = u_interpolant.value(p); }
else if (component == 1) { res = T_interpolant.value(p); }
else if (component == 2) { res = lambda_interpolant.value(p); }
return res;
}
template class SolutionVectorFunction<Interpolant>;
}
Annotated version of Solution.h
#ifndef SOLUTION
#define SOLUTION
#include <deal.II/base/function.h>
#include "LinearInterpolator.h"
#include <vector>
#include <string>
#include <fstream>
#include <iostream>
#include <iomanip>
namespace TravelingWave
{
The structure for keeping the solution: arrays of coordinates \xi, solution u, T, \lambda, and the wave speed c.
struct SolutionStruct
{
SolutionStruct();
SolutionStruct(const std::vector<double> &ix, const std::vector<double> &iu,
const std::vector<double> &iT, const std::vector<double> &ilambda, const double iwave_speed);
SolutionStruct(const std::vector<double> &ix, const std::vector<double> &iu,
const std::vector<double> &iT, const std::vector<double> &ilambda);
void reinit(const unsigned int number_of_elements);
void save_to_file(std::string filename) const;
std::vector<double> x;
std::vector<double> u;
std::vector<double> T;
std::vector<double> lambda;
double wave_speed;
};
Interpolation class
{
public:
Interpolant(const std::vector<double> &ix_points, const std::vector<double> &iy_points);
virtual double value(
const Point<1> &p,
const unsigned int component = 0)
const override;
private:
LinearInterpolator<double> interpolant;
};
virtual RangeNumberType value(const Point< dim > &p, const unsigned int component=0) const
Vector function (u(p), T(p), \lambda(p))
template <typename InterpolantType>
class SolutionVectorFunction :
public Function<1>
{
public:
SolutionVectorFunction(InterpolantType iu_interpolant, InterpolantType iT_interpolant, InterpolantType ilambda_interpolant);
virtual double value(
const Point<1> &p,
const unsigned int component = 0)
const override;
private:
InterpolantType u_interpolant;
InterpolantType T_interpolant;
InterpolantType lambda_interpolant;
};
}
#endif
Annotated version of TravelingWaveSolver.cc
#include "TravelingWaveSolver.h"
namespace TravelingWave
{
Constructor of the class that takes parameters of the problem and an initial guess for Newton's iterations.
TravelingWaveSolver::TravelingWaveSolver(const Parameters ¶meters, const SolutionStruct &initial_guess_input)
: params(parameters)
, problem(params.problem)
, number_of_quadrature_points((params.fe.quadrature_points_number > 0) ? params.fe.quadrature_points_number : (params.fe.poly_degree + 1))
, triangulation_uploaded(false)
, fe(
FE_Q<1>(params.fe.poly_degree), 1,
FE_Q<1>(params.fe.poly_degree), 1,
FE_Q<1>(params.fe.poly_degree), 1)
, current_wave_speed(0.)
, initial_guess(initial_guess_input)
{
const ::parallel::distributed::Triangulation< dim, spacedim > * triangulation
Table with values of some parameters to be written to the standard output before calculations.
table.
add_value(
"Parameter name",
"number of quadrature points");
table.add_value("value", number_of_quadrature_points);
table.add_value("Parameter name", "delta");
table.add_value("value", params.problem.delta);
table.add_value("Parameter name", "epsilon");
table.add_value("value", params.problem.epsilon);
table.add_value("Parameter name", "params.problem.wave_speed_init");
table.add_value("value", params.problem.wave_speed_init);
table.add_value("Parameter name", "initial_guess.wave_speed");
table.add_value("value", initial_guess.wave_speed);
table.add_value("Parameter name", "T_left");
table.add_value("value", params.problem.T_left);
table.set_precision("value", 2);
table.set_scientific("value", true);
std::cout << "\n";
std::cout << "\n";
}
void add_value(const std::string &key, const T value)
A function that takes a triangulation and assigns it to the member variable triangulation .
void TravelingWaveSolver::set_triangulation(
const Triangulation<1> &itriangulation)
{
triangulation_uploaded = true;
}
virtual void copy_triangulation(const ::Triangulation< dim, spacedim > &other_tria) override
virtual void clear() override
Here we find the indices of the degrees of freedom, associated with the boundary vertices, and the degree of freedom, associated with the vertex with coordinate \xi = 0, and corresponding to temperature.
void TravelingWaveSolver::find_boundary_and_centering_dof_numbers()
{
for (const auto &cell : dof_handler.active_cell_iterators())
{
{
if (isapprox(cell->vertex(v_ind)[0], params.mesh.interval_left))
{
boundary_and_centering_dof_numbers["u_left"] = cell->vertex_dof_index(v_ind, 0);
boundary_and_centering_dof_numbers["T_left"] = cell->vertex_dof_index(v_ind, 1);
boundary_and_centering_dof_numbers["lambda_left"] = cell->vertex_dof_index(v_ind, 2);
}
else if (isapprox(cell->vertex(v_ind)[0], params.mesh.interval_right))
{
boundary_and_centering_dof_numbers["u_right"] = cell->vertex_dof_index(v_ind, 0);
boundary_and_centering_dof_numbers["T_right"] = cell->vertex_dof_index(v_ind, 1);
boundary_and_centering_dof_numbers["lambda_right"] = cell->vertex_dof_index(v_ind, 2);
}
else if (isapprox(cell->vertex(v_ind)[0], 0.))
{
boundary_and_centering_dof_numbers["T_zero"] = cell->vertex_dof_index(v_ind, 1);
}
}
}
}
Set solution values, corresponding to Dirichlet boundary conditions and the centering condition T(0) = T_{\mathrm{ign}}.
void TravelingWaveSolver::set_boundary_and_centering_values()
{
current_solution[boundary_and_centering_dof_numbers["u_right"]] = problem.u_right;
current_solution[boundary_and_centering_dof_numbers["T_left"]] = problem.T_left;
if (problem.T_r_bc_type == 1)
{
current_solution[boundary_and_centering_dof_numbers["T_right"]] = problem.T_right;
}
current_solution[boundary_and_centering_dof_numbers["T_zero"]] = problem.T_ign;
current_solution[boundary_and_centering_dof_numbers["lambda_right"]] = 0.;
}
void TravelingWaveSolver::setup_system(const bool initial_step)
{
dof_handler.distribute_dofs(fe);
std::cout << "Number of dofs : " << dof_handler.n_dofs() << std::endl;
extended_solution_dim = dof_handler.n_dofs() + 1;
find_boundary_and_centering_dof_numbers();
Boundary condition constraints for du, dT and d\lambda.
zero_boundary_constraints.clear();
Dirichlet homogeneous boundary condition for du at the right boundary.
zero_boundary_constraints.add_line(boundary_and_centering_dof_numbers["u_right"]);
Dirichlet homogeneous boundary condition for dT at the left boundary.
zero_boundary_constraints.add_line(boundary_and_centering_dof_numbers["T_left"]);
For the temperature at the left boundary there are two possibilities:
if (problem.T_r_bc_type == 1)
{
std::cout << "Dirichlet condition for the temperature at the right boundary." << std::endl;
zero_boundary_constraints.add_line(boundary_and_centering_dof_numbers["T_right"]);
}
else
{
std::cout << "Neumann condition for the temperature at the right boundary." << std::endl;
}
Dirichlet homogeneous boundary condition for d\lambda at the right boundary. (At the left boundary we consider the homogeneous Neumann boundary condition for d\lambda.)
zero_boundary_constraints.add_line(boundary_and_centering_dof_numbers["lambda_right"]);
zero_boundary_constraints.close();
We create extended dynamic sparsity pattern with an additional row and an additional column.
{
std::vector<types::global_dof_index> dofs_on_this_cell;
dofs_on_this_cell.reserve(dof_handler.get_fe_collection().max_dofs_per_cell());
for (const auto &cell : dof_handler.active_cell_iterators())
{
const unsigned
int dofs_per_cell = cell->get_fe().n_dofs_per_cell();
dofs_on_this_cell.resize(dofs_per_cell);
cell->get_dof_indices(dofs_on_this_cell);
zero_boundary_constraints.add_entries_local_to_global(dofs_on_this_cell,
dsp,
true);
}
Adding elements to the last column.
for (unsigned int i = 0; i < extended_solution_dim; ++i)
{
dsp.add(i, extended_solution_dim - 1);
}
Adding one element to the last row, corresponding to the T(0).
dsp.add(extended_solution_dim - 1, boundary_and_centering_dof_numbers["T_zero"]);
}
Initialization
sparsity_pattern_extended.copy_from(dsp);
jacobian_matrix_extended.reinit(sparsity_pattern_extended);
jacobian_matrix_extended_factorization.reset();
current_solution_extended.reinit(extended_solution_dim);
if (initial_step)
{
current_solution.reinit(dof_handler.n_dofs());
}
}
void TravelingWaveSolver::set_initial_guess()
{
current_wave_speed = initial_guess.wave_speed;
The initial condition is a discrete set of coordinates \xi and values of functions u, T and \lambda. From the three sets we create three continuous functions using interpolation, which then form one continuous vector function of SolutionVectorFunction type.
Interpolant u_interpolant(initial_guess.x, initial_guess.u);
Interpolant T_interpolant(initial_guess.x, initial_guess.T);
Interpolant lambda_interpolant(initial_guess.x, initial_guess.lambda);
SolutionVectorFunction init_guess_func(u_interpolant, T_interpolant, lambda_interpolant);
set_boundary_and_centering_values();
for (unsigned int i = 0; i < extended_solution_dim - 1; ++i)
{
current_solution_extended(i) = current_solution(i);
}
current_solution_extended(extended_solution_dim - 1) = current_wave_speed;
}
Heaviside function.
double TravelingWaveSolver::Heaviside_func(double x) const
{
if (x > 0)
{
return 1.;
}
else
{
return 0.;
}
}
void TravelingWaveSolver::compute_and_factorize_jacobian(
const Vector<double> &evaluation_point_extended)
{
{
for (unsigned int i = 0; i < dof_handler.n_dofs(); ++i)
{
evaluation_point(i) = evaluation_point_extended(i);
}
const double wave_speed = evaluation_point_extended(extended_solution_dim - 1);
std::cout << "Computing Jacobian matrix ... " << std::endl;
const QGauss<1> quadrature_formula(number_of_quadrature_points);
jacobian_matrix_extended = 0;
quadrature_formula,
const unsigned int dofs_per_cell = fe.n_dofs_per_cell();
const unsigned int n_q_points = quadrature_formula.size();
std::vector<types::global_dof_index> local_dof_indices(dofs_per_cell);
std::vector<double> current_velocity_values(n_q_points);
std::vector<double> current_temperature_values(n_q_points);
std::vector<double> current_lambda_values(n_q_points);
std::vector<Tensor<1, 1>> current_velocity_gradients(n_q_points);
std::vector<Tensor<1, 1>> current_temperature_gradients(n_q_points);
std::vector<Tensor<1, 1>> current_lambda_gradients(n_q_points);
std::vector<double> phi_u(dofs_per_cell);
std::vector<Tensor<1, 1>> grad_phi_u(dofs_per_cell);
std::vector<double> phi_T(dofs_per_cell);
std::vector<Tensor<1, 1>> grad_phi_T(dofs_per_cell);
std::vector<double> phi_lambda(dofs_per_cell);
std::vector<Tensor<1, 1>> grad_phi_lambda(dofs_per_cell);
for (const auto &cell : dof_handler.active_cell_iterators())
{
row_last_element_vector = 0;
fe_values.reinit(cell);
fe_values[velocity].get_function_values(evaluation_point, current_velocity_values);
fe_values[temperature].get_function_values(evaluation_point, current_temperature_values);
fe_values[
lambda].get_function_values(evaluation_point, current_lambda_values);
fe_values[velocity].get_function_gradients(evaluation_point, current_velocity_gradients);
fe_values[temperature].get_function_gradients(evaluation_point, current_temperature_gradients);
fe_values[
lambda].get_function_gradients(evaluation_point, current_lambda_gradients);
auto kappa_1 = [=](
double T,
double lambda){
problem.theta / (T * T) * Heaviside_func(T - problem.T_ign)
);
};
auto kappa_2 = [=](
double T,
double lambda){
return -problem.k *
std::exp(-problem.theta / T) * Heaviside_func(T - problem.T_ign);
};
for (unsigned int q = 0; q < n_q_points; ++q)
{
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
phi_u[k] = fe_values[velocity].value(k, q);
grad_phi_u[k] = fe_values[velocity].gradient(k, q);
phi_T[k] = fe_values[temperature].value(k, q);
grad_phi_T[k] = fe_values[temperature].gradient(k, q);
phi_lambda[k] = fe_values[
lambda].value(k, q);
grad_phi_lambda[k] = fe_values[
lambda].gradient(k, q);
}
const double del_Pr_eps = (problem.Pr * 4 * problem.delta / (3 * problem.epsilon));
const double del_Le = (problem.delta / problem.Le);
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
for (unsigned int j = 0; j < dofs_per_cell; ++j)
{
del_Pr_eps * (-grad_phi_u[i] * grad_phi_u[j])
+ phi_u[i] * (
- (1 - wave_speed + problem.epsilon * current_velocity_values[q]) * grad_phi_u[j][0]
- problem.epsilon * current_velocity_gradients[q][0] * phi_u[j]
- problem.epsilon / 2. * grad_phi_T[j][0]
)
+ problem.delta * (-grad_phi_T[i] * grad_phi_T[j])
+ phi_T[i] * (
- wave_speed * grad_phi_u[j][0]
+ wave_speed * grad_phi_T[j][0]
+ problem.q * kappa_1(current_temperature_values[q], current_lambda_values[q]) * phi_T[j]
+ problem.q * kappa_2(current_temperature_values[q], current_lambda_values[q]) * phi_lambda[j]
)
+ del_Le * (-grad_phi_lambda[i] * grad_phi_lambda[j])
+ phi_lambda[i] * (
kappa_1(current_temperature_values[q], current_lambda_values[q]) * phi_T[j]
+ wave_speed * grad_phi_lambda[j][0]
+ kappa_2(current_temperature_values[q], current_lambda_values[q]) * phi_lambda[j]
)
) * fe_values.JxW(q);
}
row_last_element_vector(i) += (
(phi_u[i] * current_velocity_gradients[q][0])
+ (phi_T[i] * current_temperature_gradients[q][0])
- (phi_T[i] * current_velocity_gradients[q][0])
+ (phi_lambda[i] * current_lambda_gradients[q][0])
) * fe_values.JxW(q);
}
}
cell->get_dof_indices(local_dof_indices);
for (const unsigned int i : fe_values.dof_indices())
{
for (const unsigned
int j : fe_values.dof_indices())
{
jacobian_matrix_extended.add(local_dof_indices[i],
local_dof_indices[j],
}
@ update_values
Shape function values.
@ update_JxW_values
Transformed quadrature weights.
@ update_gradients
Shape function gradients.
@ update_quadrature_points
Transformed quadrature points.
void cell_matrix(FullMatrix< double > &M, const FEValuesBase< dim > &fe, const FEValuesBase< dim > &fetest, const ArrayView< const std::vector< double > > &velocity, const double factor=1.)
Adding elements to the last column.
jacobian_matrix_extended.add(local_dof_indices[i],
extended_solution_dim - 1,
row_last_element_vector(i));
}
}
Global dof indices of dofs for dT and d\lambda, associated with vertex \xi = 0.
Approximating the derivative of T at \xi = 0 as done in step-14.
double T_point_derivative(0.);
double T_at_zero_point(0.);
double lambda_at_zero_point(0.);
{
double derivative_evaluation_point = 0.;
quadrature_formula,
const unsigned int n_q_points = quadrature_formula.size();
std::vector<double> current_temperature_values(n_q_points);
std::vector<Tensor<1, 1>> current_temperature_gradients(n_q_points);
std::vector<double> current_lambda_values(n_q_points);
unsigned int derivative_evaluation_point_hits = 0;
for (const auto &cell : dof_handler.active_cell_iterators())
{
{
if (isapprox(cell->vertex(vertex)[0], derivative_evaluation_point))
{
T_zero_point_dof_ind = cell->vertex_dof_index(vertex, 1);
lambda_zero_point_dof_ind = cell->vertex_dof_index(vertex, 2);
fe_values.reinit(cell);
fe_values[temperature].get_function_values(current_solution, current_temperature_values);
fe_values[temperature].get_function_gradients(current_solution, current_temperature_gradients);
fe_values[lambda].get_function_values(current_solution, current_lambda_values);
unsigned int q_point = 0;
for (; q_point < n_q_points; ++q_point)
{
if (isapprox(fe_values.quadrature_point(q_point)[0], derivative_evaluation_point))
{
break;
}
}
T_at_zero_point = current_temperature_values[q_point];
lambda_at_zero_point = current_lambda_values[q_point];
T_point_derivative += current_temperature_gradients[q_point][0];
++derivative_evaluation_point_hits;
break;
}
}
}
T_point_derivative /= static_cast<double>(derivative_evaluation_point_hits);
}
Here we add to the matrix the terms that appear after integrating the terms with the Dirac delta function (which we skipped inside the loop).
double term_with_delta_func(0.);
term_with_delta_func = problem.k *
std::exp(-problem.theta / T_at_zero_point) * (1 - lambda_at_zero_point) /
std::abs(T_point_derivative);
jacobian_matrix_extended.add(T_zero_point_dof_ind, T_zero_point_dof_ind, problem.q * term_with_delta_func);
jacobian_matrix_extended.add(lambda_zero_point_dof_ind, T_zero_point_dof_ind, term_with_delta_func);
Add 1 to the position T_zero_point_dof_ind of the last row of the matrix.
jacobian_matrix_extended.add(extended_solution_dim - 1, T_zero_point_dof_ind, 1.);
zero_boundary_constraints.condense(jacobian_matrix_extended);
}
{
std::cout << "Factorizing Jacobian matrix" << std::endl;
jacobian_matrix_extended_factorization = std::make_unique<SparseDirectUMFPACK>();
jacobian_matrix_extended_factorization->factorize(jacobian_matrix_extended);
}
}
{
std::cout << "Computing residual vector ... " << std::endl;
residual = 0;
for (unsigned int i = 0; i < dof_handler.n_dofs(); ++i)
{
evaluation_point(i) = evaluation_point_extended(i);
}
const double wave_speed = evaluation_point_extended(extended_solution_dim - 1);
const QGauss<1> quadrature_formula(number_of_quadrature_points);
quadrature_formula,
const unsigned int dofs_per_cell = fe.n_dofs_per_cell();
const unsigned int n_q_points = quadrature_formula.size();
std::vector<types::global_dof_index> local_dof_indices(dofs_per_cell);
std::vector<double> current_velocity_values(n_q_points);
std::vector<Tensor<1, 1>> current_velocity_gradients(n_q_points);
std::vector<double> current_temperature_values(n_q_points);
std::vector<Tensor<1, 1>> current_temperature_gradients(n_q_points);
std::vector<double> current_lambda_values(n_q_points);
std::vector<Tensor<1, 1>> current_lambda_gradients(n_q_points);
std::vector<double> phi_u(dofs_per_cell);
std::vector<Tensor<1, 1>> grad_phi_u(dofs_per_cell);
std::vector<double> phi_T(dofs_per_cell);
std::vector<Tensor<1, 1>> grad_phi_T(dofs_per_cell);
std::vector<double> phi_lambda(dofs_per_cell);
std::vector<Tensor<1, 1>> grad_phi_lambda(dofs_per_cell);
for (const auto &cell : dof_handler.active_cell_iterators())
{
fe_values.reinit(cell);
fe_values[velocity].get_function_values(evaluation_point, current_velocity_values);
fe_values[velocity].get_function_gradients(evaluation_point, current_velocity_gradients);
fe_values[temperature].get_function_values(evaluation_point, current_temperature_values);
fe_values[temperature].get_function_gradients(evaluation_point, current_temperature_gradients);
fe_values[
lambda].get_function_values(evaluation_point, current_lambda_values);
fe_values[
lambda].get_function_gradients(evaluation_point, current_lambda_gradients);
auto omega = [=](
double T,
double lambda){
return problem.k * (1 -
lambda) *
std::exp(-problem.theta / T) * Heaviside_func(T - problem.T_ign);
};
for (unsigned int q = 0; q < n_q_points; ++q)
{
for (unsigned int k = 0; k < dofs_per_cell; ++k)
{
phi_u[k] = fe_values[velocity].value(k, q);
grad_phi_u[k] = fe_values[velocity].gradient(k, q);
phi_T[k] = fe_values[temperature].value(k, q);
grad_phi_T[k] = fe_values[temperature].gradient(k, q);
phi_lambda[k] = fe_values[
lambda].value(k, q);
grad_phi_lambda[k] = fe_values[
lambda].gradient(k, q);
}
double del_Pr_eps = (problem.Pr * 4 * problem.delta / (3 * problem.epsilon));
double del_Le = (problem.delta / problem.Le);
for (unsigned int i = 0; i < dofs_per_cell; ++i)
{
del_Pr_eps * (-grad_phi_u[i] * current_velocity_gradients[q])
+ phi_u[i] * (
- current_velocity_gradients[q][0] * (1 - wave_speed + problem.epsilon * current_velocity_values[q])
- problem.epsilon / 2. * current_temperature_gradients[q][0]
)
+ problem.delta * (-grad_phi_T[i] * current_temperature_gradients[q])
+ phi_T[i] * (
wave_speed * (current_temperature_gradients[q][0] - current_velocity_gradients[q][0])
+ problem.q * omega(current_temperature_values[q], current_lambda_values[q])
)
+ del_Le * (-grad_phi_lambda[i] * current_lambda_gradients[q])
+ phi_lambda[i] * (
wave_speed * current_lambda_gradients[q][0] + omega(current_temperature_values[q], current_lambda_values[q])
)
) * fe_values.JxW(q);
}
}
cell->get_dof_indices(local_dof_indices);
for (const unsigned int i : fe_values.dof_indices())
{
}
}
residual(extended_solution_dim - 1) = 0.;
zero_boundary_constraints.condense(residual);
double residual_norm = residual.
l2_norm();
std::cout << std::defaultfloat;
std::cout << "norm of residual = " << residual_norm << std::endl;
return residual_norm;
}
real_type l2_norm() const
void cell_residual(Vector< double > &result, const FEValuesBase< dim > &fe, const std::vector< Tensor< 1, dim > > &input, const ArrayView< const std::vector< double > > &velocity, double factor=1.)
Split the solution vector into two parts: one part is the solution u, T and \lambda, and another part is the wave speed.
void TravelingWaveSolver::split_extended_solution_vector()
{
for (unsigned int i = 0; i < extended_solution_dim - 1; ++i)
{
current_solution(i) = current_solution_extended(i);
}
current_wave_speed = current_solution_extended(extended_solution_dim - 1);
}
{
std::cout << "Solving linear system ... " << std::endl;
jacobian_matrix_extended_factorization->vmult(solution_extended, rhs);
zero_boundary_constraints.distribute(solution_extended);
}
Function for adaptive mesh refinement based on KellyErrorEstimator .
void TravelingWaveSolver::refine_mesh()
{
dof_handler,
{},
current_solution,
estimated_error_per_cell,
fe.component_mask(lambda)
);
estimated_error_per_cell,
0.1,
0.05);
solution_transfer.prepare_for_coarsening_and_refinement(current_solution);
setup_system( false);
solution_transfer.interpolate(current_solution, tmp);
current_solution = std::move(tmp);
set_boundary_and_centering_values();
for (unsigned int i = 0; i < extended_solution_dim - 1; ++i)
{
current_solution_extended(i) = current_solution(i);
}
current_solution_extended(extended_solution_dim - 1) = current_wave_speed;
}
double TravelingWaveSolver::run_newton_iterations(const double target_tolerance)
{
double residual_norm = 0.;
{
additional_data.function_tolerance = target_tolerance;
x.reinit(extended_solution_dim);
};
residual_norm = compute_residual(evaluation_point, residual);
return 0;
};
compute_and_factorize_jacobian(evaluation_point);
return 0;
};
this->solve(rhs, solution, tolerance);
return 0;
};
nonlinear_solver.solve(current_solution_extended);
}
return residual_norm;
}
static void estimate(const Mapping< dim, spacedim > &mapping, const DoFHandler< dim, spacedim > &dof, const Quadrature< dim - 1 > &quadrature, const std::map< types::boundary_id, const Function< spacedim, Number > * > &neumann_bc, const ReadVector< Number > &solution, Vector< float > &error, const ComponentMask &component_mask={}, const Function< spacedim > *coefficients=nullptr, const unsigned int n_threads=numbers::invalid_unsigned_int, const types::subdomain_id subdomain_id=numbers::invalid_subdomain_id, const types::material_id material_id=numbers::invalid_material_id, const Strategy strategy=cell_diameter_over_24)
unsigned int n_active_cells() const
virtual void execute_coarsening_and_refinement() override
virtual bool prepare_coarsening_and_refinement() override
void refine_and_coarsen_fixed_number(Triangulation< dim, spacedim > &triangulation, const Vector< Number > &criteria, const double top_fraction_of_cells, const double bottom_fraction_of_cells, const unsigned int max_n_cells=std::numeric_limits< unsigned int >::max())
Output the solution ( u, T and \lambda) and the wave speed into two separate files with double precision. The files can be read by gnuplot.
void TravelingWaveSolver::output_with_double_precision(
const Vector<double> &solution,
const double wave_speed,
const std::string filename)
{
const std::string file_for_solution = filename + ".txt";
std::ofstream output(file_for_solution);
for (const auto &cell : dof_handler.active_cell_iterators())
{
{
double u = solution(cell->vertex_dof_index(v_ind, 0));
double T = solution(cell->vertex_dof_index(v_ind, 1));
double lambda = solution(cell->vertex_dof_index(v_ind, 2));
output << std::scientific << std::setprecision(16);
output << cell->vertex(v_ind)[0];
output << std::scientific << std::setprecision(16);
output << std::scientific << " " << u << " " << T << " " << lambda << "\n";
}
output << "\n";
}
output.close();
std::ofstream file_for_wave_speed_output("wave_speed-" + file_for_solution);
file_for_wave_speed_output << std::scientific << std::setprecision(16);
file_for_wave_speed_output << wave_speed << std::endl;
file_for_wave_speed_output.close();
}
Copy the solution into the SolutionStruct object, that stores the solution in an ordered manner.
void TravelingWaveSolver::get_solution(SolutionStruct &solution) const
{
To obtain an ordered solution array, we first create a set consisting of the elements {x, u, T, lambda} in which the sorting is done by coordinate, and then copy the contents of the set into the arrays of the SolutionStruct object.
auto comp = [](const std::vector<double> &a, const std::vector<double> &b) {
return a[0] < b[0];
};
std::set<std::vector<double>, decltype(comp)> solution_set(comp);
for (const auto &cell : dof_handler.active_cell_iterators())
{
{
double x = cell->vertex(v_ind)[0];
double u = current_solution(cell->vertex_dof_index(v_ind, 0));
double T = current_solution(cell->vertex_dof_index(v_ind, 1));
double lambda = current_solution(cell->vertex_dof_index(v_ind, 2));
solution_set.insert({x, u, T, lambda});
}
}
solution.x.clear();
solution.u.clear();
solution.T.clear();
solution.lambda.clear();
solution.x.reserve(solution_set.size());
solution.u.reserve(solution_set.size());
solution.T.reserve(solution_set.size());
solution.lambda.reserve(solution_set.size());
for (auto it = solution_set.begin(); it != solution_set.end(); ++it)
{
solution.x.push_back((*it)[0]);
solution.u.push_back((*it)[1]);
solution.T.push_back((*it)[2]);
solution.lambda.push_back((*it)[3]);
}
solution.wave_speed = current_wave_speed;
}
void TravelingWaveSolver::get_triangulation(
Triangulation<1> &otriangulation)
const
{
otriangulation.clear();
}
void TravelingWaveSolver::run(const std::string filename, const bool save_solution_to_file)
{
const int mesh_refinement_type = params.mesh.adaptive;
const unsigned int n_refinements = params.mesh.refinements_number;
const double tol = params.solver.tol;
if (triangulation_uploaded == false)
{
We create two triangulations: one to the left and one to the right of zero coordinate. After that we merge them to obtain one triangulation, which contains zero point.
triangulation_left,
static_cast<unsigned int>(
std::abs( 0. - params.mesh.interval_left )),
params.mesh.interval_left, 0.
);
triangulation_right,
static_cast<unsigned int>(
std::abs( params.mesh.interval_right - 0. )),
0., params.mesh.interval_right
);
}
if (triangulation_uploaded == false)
{
if (mesh_refinement_type == 1)
{
}
else if (mesh_refinement_type == 0)
{
}
}
setup_system( true);
set_initial_guess();
if (save_solution_to_file)
{
output_with_double_precision(current_solution, current_wave_speed, "solution_initial_data");
}
if (mesh_refinement_type == 1)
{
double residual_norm = 0.;
{
residual_norm = compute_residual(current_solution_extended, tmp_residual);
}
unsigned int refinement_cycle = 0;
while ((residual_norm > tol) && (refinement_cycle < n_refinements))
{
computing_timer.reset();
std::cout << "Mesh refinement step " << refinement_cycle << std::endl;
if (refinement_cycle != 0) { refine_mesh(); }
const double target_tolerance = 0.1 *
std::pow(0.1, refinement_cycle);
std::cout << " Target_tolerance: " << target_tolerance << std::endl;
residual_norm = run_newton_iterations(target_tolerance);
split_extended_solution_vector();
{
std::cout << std::scientific << std::setprecision(16);
std::cout << "current_wave_speed = " << current_wave_speed << std::endl;
std::cout << std::defaultfloat;
}
computing_timer.print_summary();
++refinement_cycle;
}
if (save_solution_to_file)
{
output_with_double_precision(current_solution, current_wave_speed, filename);
}
}
else if (mesh_refinement_type == 0)
{
run_newton_iterations(tol);
split_extended_solution_vector();
if (save_solution_to_file)
{
output_with_double_precision(current_solution, current_wave_speed, filename);
}
{
std::cout << std::scientific << std::setprecision(16);
std::cout << "current_wave_speed = " << current_wave_speed << std::endl;
std::cout << std::defaultfloat;
}
computing_timer.print_summary();
}
}
}
void refine_global(const unsigned int times=1)
void subdivided_hyper_cube(Triangulation< dim, spacedim > &tria, const unsigned int repetitions, const double left=0., const double right=1., const bool colorize=false)
void merge_triangulations(const Triangulation< dim, spacedim > &triangulation_1, const Triangulation< dim, spacedim > &triangulation_2, Triangulation< dim, spacedim > &result, const double duplicated_vertex_tolerance=1.0e-12, const bool copy_manifold_ids=false, const bool copy_boundary_ids=false)
::VectorizedArray< Number, width > pow(const ::VectorizedArray< Number, width > &, const Number p)
Annotated version of TravelingWaveSolver.h
#ifndef TRAVELING_WAVE_SOLVER
#define TRAVELING_WAVE_SOLVER
#include <deal.II/base/timer.h>
#include <deal.II/base/function.h>
#include <deal.II/base/table_handler.h>
#include <deal.II/grid/tria.h>
#include <deal.II/grid/grid_generator.h>
#include <deal.II/grid/grid_refinement.h>
#include <deal.II/grid/grid_out.h>
#include <deal.II/dofs/dof_handler.h>
#include <deal.II/dofs/dof_tools.h>
#include <deal.II/fe/fe_q.h>
#include <deal.II/fe/fe_system.h>
#include <deal.II/fe/fe_values.h>
#include <deal.II/lac/vector.h>
#include <deal.II/lac/full_matrix.h>
#include <deal.II/lac/sparse_matrix.h>
#include <deal.II/lac/sparse_direct.h>
#include <deal.II/lac/dynamic_sparsity_pattern.h>
#include <deal.II/numerics/vector_tools.h>
#include <deal.II/numerics/error_estimator.h>
#include <deal.II/numerics/solution_transfer.h>
#include <deal.II/numerics/data_out.h>
#include <deal.II/sundials/kinsol.h>
#include "Parameters.h"
#include "Solution.h"
#include "AuxiliaryFunctions.h"
#include <cmath>
#include <algorithm>
#include <string>
#include <fstream>
#include <iostream>
#include <iomanip>
#include <set>
Namespace of the program
namespace TravelingWave
{
The main class for construction of the traveling wave solutions.
class TravelingWaveSolver
{
public:
TravelingWaveSolver(const Parameters ¶meters, const SolutionStruct &initial_guess_input);
void run(const std::string filename="solution", const bool save_solution_to_file=true);
void get_solution(SolutionStruct &solution) const;
private:
void setup_system(const bool initial_step);
void find_boundary_and_centering_dof_numbers();
void set_boundary_and_centering_values();
void set_initial_guess();
double Heaviside_func(double x) const;
void compute_and_factorize_jacobian(
const Vector<double> &evaluation_point_extended);
void split_extended_solution_vector();
void refine_mesh();
double run_newton_iterations(const double target_tolerance=1e-5);
void output_with_double_precision(
const Vector<double> &solution,
const double wave_speed,
const std::string filename=
"solution");
The dimension of the finite element solution increased by one to account for the value corresponding to the wave speed.
unsigned int extended_solution_dim;
std::map<std::string, unsigned int> boundary_and_centering_dof_numbers;
Parameters of the problem, taken from a .prm file.
const Parameters ¶ms;
const Problem &problem;
unsigned int number_of_quadrature_points;
The flag indicating whether the triangulation was uploaded externally or created within the run member function.
bool triangulation_uploaded;
Constraints for Dirichlet boundary conditions.
std::unique_ptr<SparseDirectUMFPACK> jacobian_matrix_extended_factorization;
Finite element solution of the problem.
Value of the wave speed c.
double current_wave_speed;
Solution with an additional term, corresponding to the variable wave_speed.
Initial guess for Newton's iterations.
SolutionStruct initial_guess;
};
}
#endif
Annotated version of calculate_profile.cc
#include "TravelingWaveSolver.h"
#include "calculate_profile.h"
namespace TravelingWave
{
Computation of the limit case (ideal) solution, corresponding to \delta = 0, by solving the ODE. The output is the part of the solution to the left of zero. Here u_0, T_0, lambda_0 are the values of the medium state to the right of zero.
void compute_limit_sol_left_part(const Parameters ¶meters,
const double wave_speed,
const double u_0,
const double T_0,
const double lambda_0,
SolutionStruct &LimitSol,
const double root_sign)
{
LimitSolution limit_sol(parameters, lambda_0, u_0, T_0, root_sign);
limit_sol.set_wave_speed(wave_speed);
{
We take more integration points to better resolve the transition layer.
std::vector<double> t_span(
static_cast<unsigned int>(
std::abs( 0. - parameters.mesh.interval_left )));
double finer_mesh_starting_value = -9.1;
linspace(parameters.mesh.interval_left, finer_mesh_starting_value, t_span);
std::vector<double> fine_grid(10000);
linspace(finer_mesh_starting_value + 1e-4, 0., fine_grid);
t_span.insert(t_span.end(), fine_grid.begin(), fine_grid.end());
Reverse the order of the elements (because we need to perform back in time integration).
std::reverse(t_span.begin(), t_span.end());
state_type lambda_val(1);
lambda_val[0] = lambda_0;
IntegrateSystemAtTimePoints(limit_sol.lambda_vec, limit_sol.t_vec, t_span,
limit_sol,
lambda_val,
-1e-2, Integrator_Type::dopri5);
}
limit_sol.calculate_u_T_omega();
Reverse the order of elements
std::reverse(limit_sol.t_vec.begin(), limit_sol.t_vec.end());
std::reverse(limit_sol.lambda_vec.begin(), limit_sol.lambda_vec.end());
std::reverse(limit_sol.u_vec.begin(), limit_sol.u_vec.end());
std::reverse(limit_sol.T_vec.begin(), limit_sol.T_vec.end());
std::reverse(limit_sol.omega_vec.begin(), limit_sol.omega_vec.end());
SaveSolutionIntoFile(limit_sol.lambda_vec, limit_sol.t_vec, "solution_lambda_limit.txt");
SaveSolutionIntoFile(limit_sol.u_vec, limit_sol.t_vec, "solution_u_limit.txt");
SaveSolutionIntoFile(limit_sol.T_vec, limit_sol.t_vec, "solution_T_limit.txt");
SaveSolutionIntoFile(limit_sol.omega_vec, limit_sol.t_vec, "solution_omega_limit.txt");
LimitSol.reinit(limit_sol.t_vec.size());
LimitSol.wave_speed = wave_speed;
for (unsigned int i=0; i < limit_sol.t_vec.size(); ++i)
{
LimitSol.x[i] = limit_sol.t_vec[i];
LimitSol.u[i] = limit_sol.u_vec[i][0];
LimitSol.T[i] = limit_sol.T_vec[i][0];
LimitSol.lambda[i] = limit_sol.lambda_vec[i][0];
}
}
Construction of an initial guess for detonation wave solution. The ODE is solved for the ideal system with \delta = 0.
void compute_initial_guess_detonation(const Parameters ¶ms, SolutionStruct &initial_guess, const double root_sign)
{
const Problem &problem = params.problem;
double current_wave_speed(problem.wave_speed_init);
{
double DeltaT = problem.T_left - problem.T_right;
double qDT = problem.q - DeltaT;
current_wave_speed = 1. + problem.epsilon * (problem.u_right - (qDT * qDT + DeltaT) / (2 * qDT));
}
double u_0 = problem.u_right;
double T_0 = problem.T_right;
double lambda_0 = 0.;
compute_limit_sol_left_part(params, current_wave_speed, u_0, T_0, lambda_0, initial_guess, root_sign);
initial_guess.wave_speed = current_wave_speed;
for (int i = initial_guess.x.size() - 1; i > - 1; --i)
{
if (isapprox(initial_guess.x[i], 0.))
{
initial_guess.u[i] = problem.u_right;
initial_guess.T[i] = problem.T_ign;
initial_guess.lambda[i] = 0.;
break;
}
}
Adding the points to the right part of the interval (w.r.t. \xi = 0).
unsigned int number_of_additional_points = 5;
for (unsigned int i = 0; i < number_of_additional_points; ++i)
{
initial_guess.x.push_back(params.mesh.interval_right / (
std::pow(2., number_of_additional_points - 1 - i)));
initial_guess.u.push_back(problem.u_right);
initial_guess.T.push_back(problem.T_right);
initial_guess.lambda.push_back(0.);
}
}
Construction of a piecewise constant initial guess for deflagration wave solution.
void compute_initial_guess_deflagration(const Parameters ¶ms, SolutionStruct &initial_guess)
{
const Problem &problem = params.problem;
double current_wave_speed(problem.wave_speed_init);
double del_Pr_eps = (problem.Pr * 4 * problem.delta / (3 * problem.epsilon));
double del_Le = (problem.delta / problem.Le);
auto u_init_guess_func = [&](double x) {
if (x < 0.)
{
return problem.u_left;
}
else
{
return problem.u_right;
}
};
auto T_init_guess_func = [&](double x) {
if (x < 0.)
{
return problem.T_left;
}
else if (isapprox(x, 0.))
{
return problem.T_ign;
}
else
{
return problem.T_right;
}
};
auto lambda_init_guess_func = [=](double x) {
if (x < 0.)
{
}
else
{
return 0.;
}
};
unsigned int multiplier_for_number_of_points = 7;
unsigned int number_of_points = multiplier_for_number_of_points *
static_cast<unsigned int>(std::trunc(
std::abs( params.mesh.interval_right - params.mesh.interval_left )));
std::vector<double> x_span(number_of_points);
linspace(params.mesh.interval_left, params.mesh.interval_right, x_span);
std::vector<double> u_init_arr(number_of_points);
std::vector<double> T_init_arr(number_of_points);
std::vector<double> lambda_init_arr(number_of_points);
for (unsigned int i = 0; i < number_of_points; ++i)
{
u_init_arr[i] = u_init_guess_func(x_span[i]);
T_init_arr[i] = T_init_guess_func(x_span[i]);
lambda_init_arr[i] = lambda_init_guess_func(x_span[i]);
}
initial_guess.x = x_span;
initial_guess.u = u_init_arr;
initial_guess.T = T_init_arr;
initial_guess.lambda = lambda_init_arr;
initial_guess.wave_speed = current_wave_speed;
}
Compute the traveling-wave profile. The continuation method can be switched on by setting the argument continuation_for_delta as true .
void calculate_profile(Parameters& parameters,
const bool continuation_for_delta ,
const double delta_start ,
const unsigned int number_of_continuation_points)
{
SolutionStruct sol;
if (parameters.problem.wave_type == 1)
{
compute_initial_guess_detonation(parameters, sol);
}
else if (parameters.problem.wave_type == 0)
{
compute_initial_guess_deflagration(parameters, sol);
}
if (continuation_for_delta == false)
{
TravelingWaveSolver wave(parameters, sol);
wave.run(filename);
wave.get_solution(sol);
}
else
{
double delta_target = parameters.problem.delta;
parameters.problem.delta = delta_start;
std::vector<double> delta_span(number_of_continuation_points);
std::string to_string(const number value, const unsigned int digits=numbers::invalid_unsigned_int)
Generate a sequence of delta values being uniformly distributed in log10 scale.
{
double delta_start_log10 = std::log10(delta_start);
double delta_target_log10 = std::log10(delta_target);
std::vector<double> delta_log_span(delta_span.size());
linspace(delta_start_log10, delta_target_log10, delta_log_span);
for (unsigned int i = 0; i < delta_span.size(); ++i)
{
delta_span[i] =
std::pow(10, delta_log_span[i]);
}
}
bool first_iter_flag = true;
for (double delta : delta_span)
{
parameters.problem.delta = delta;
TravelingWaveSolver wave(parameters, sol);
if (first_iter_flag)
{
first_iter_flag = false;
}
else
{
wave.set_triangulation(refined_triangulation);
}
wave.run(filename);
wave.get_solution(sol);
wave.get_triangulation(refined_triangulation);
}
}
Error estimation.
{
unsigned int sol_length = sol.x.size();
double u_r = sol.u[sol_length-1];
double T_r = sol.T[sol_length-1];
double u_l = sol.u[0];
double T_l = sol.T[0];
double wave_speed = sol.wave_speed;
std::cout << "Error estimates:" << std::endl;
double DeltaT = T_l - T_r;
double qDT = parameters.problem.q - DeltaT;
double wave_speed_formula = 1. + parameters.problem.epsilon * (u_r - (qDT * qDT + DeltaT) / (2 * qDT));
std::cout << std::setw(18) << std::left << "For wave speed" << " : " << std::setw(5) << wave_speed - wave_speed_formula << std::endl;
double u_l_formula = DeltaT + u_r - parameters.problem.q;
std::cout << std::setw(18) << std::left << "For u_l" << " : " << std::setw(5) << u_l - u_l_formula << std::endl;
}
}
}
Annotated version of calculate_profile.h
#ifndef CALCULATE_PROFILE
#define CALCULATE_PROFILE
#include "Parameters.h"
#include "Solution.h"
#include "LimitSolution.h"
#include "IntegrateSystem.h"
#include "AuxiliaryFunctions.h"
namespace TravelingWave
{
void compute_limit_sol_left_part(const Parameters ¶meters,
const double wave_speed,
const double u_0,
const double T_0,
const double lambda_0,
SolutionStruct &LimitSol,
const double root_sign = 1.);
void compute_initial_guess_detonation(const Parameters ¶ms, SolutionStruct &initial_guess, const double root_sign = 1.);
void compute_initial_guess_deflagration(const Parameters ¶ms, SolutionStruct &initial_guess);
void calculate_profile(Parameters& parameters,
const bool continuation_for_delta=false ,
const double delta_start=0.01 ,
const unsigned int number_of_continuation_points=10);
}
#endif
Annotated version of main.cc
#include "calculate_profile.h"
int main(int argc, char *argv[])
{
try
{
using namespace TravelingWave;
Parameters parameters;
std::string prm_filename = "ParametersList.prm";
if (argc > 1)
{
Check if file argv[1] exists.
if (file_exists(argv[1]))
{
prm_filename = argv[1];
}
else
{
std::string errorMessage = "File \"" + std::string(argv[1]) + "\" is not found.";
throw std::runtime_error(errorMessage);
}
}
else
{
Check if the file "ParametersList.prm" exists in the current or in the parent directory.
if (!(file_exists(prm_filename) || file_exists("../" + prm_filename)))
{
std::string errorMessage = "File \"" + prm_filename + "\" is not found.";
throw std::runtime_error(errorMessage);
}
else
{
if (!file_exists(prm_filename))
{
prm_filename = "../" + prm_filename;
}
}
}
std::cout << "Reading parameters... " << std::flush;
std::cout << "done" << std::endl;
calculate_profile(parameters, false, 0.1, 3);
}
catch (std::exception &exc)
{
std::cerr << std::endl
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Exception on processing: " << std::endl
<< exc.what() << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
catch (...)
{
std::cerr << std::endl
<< std::endl
<< "----------------------------------------------------"
<< std::endl;
std::cerr << "Unknown exception!" << std::endl
<< "Aborting!" << std::endl
<< "----------------------------------------------------"
<< std::endl;
return 1;
}
return 0;
}
static void initialize(const std::string &filename="", const std::string &output_filename="", const ParameterHandler::OutputStyle output_style_for_output_filename=ParameterHandler::Short, ParameterHandler &prm=ParameterAcceptor::prm, const ParameterHandler::OutputStyle output_style_for_filename=ParameterHandler::DefaultStyle)
Annotated version of plot.py
import numpy as np
import matplotlib.pyplot as plt
import os
import sys
plot_params = {
'scatter.marker' : 'x',
'lines.markersize' : 4,
'lines.linewidth' : 1,
'axes.labelsize': 16,
'font.size' : 16,
'legend.fontsize': 16,
'xtick.labelsize': 14,
'ytick.labelsize': 14,
'text.usetex': True,
'figure.figsize': [9,6],
'axes.grid': True
}
plt.rcParams.update(plot_params)
if len(sys.argv) > 1:
filename = sys.argv[1]
if os.path.exists(filename):
data = np.loadtxt(filename, np.float64)
data_unique = np.unique(data, axis=0)
data_unique = np.array(sorted(data_unique, key=lambda x : x[0]))
x = data_unique[:, 0]
u_sol = data_unique[:, 1]
T_sol = data_unique[:, 2]
lambda_sol = data_unique[:, 3]
fig, ax = plt.subplots(nrows=1, ncols=1)
ax.scatter(x, u_sol, label=r"@f$u@f$", color='blue')
ax.scatter(x, T_sol, label=r"@f$T@f$", color='red')
ax.scatter(x, lambda_sol, label=r"@f$\lambda@f$", color='green')
'''
path_to_solution_files = os.path.split(filename)[0]
u_limit_path = os.path.join(path_to_solution_files, 'solution_u_limit.txt')
T_limit_path = os.path.join(path_to_solution_files, 'solution_T_limit.txt')
lambda_limit_path = os.path.join(path_to_solution_files, 'solution_lambda_limit.txt')
if os.path.exists(u_limit_path):
u_limit = np.loadtxt(u_limit_path, np.float64)
ax.plot(u_limit[:, 0], u_limit[:, 1], label=r"@f$u_{\mathrm{lim}}@f$", color='blue')
ax.plot([0, x[-1]], [u_sol[-1], u_sol[-1]], color='blue')
else:
print("No such file:", u_limit_path)
if os.path.exists(T_limit_path):
T_limit = np.loadtxt(T_limit_path, np.float64)
ax.plot(T_limit[:, 0], T_limit[:, 1], label=r"@f$T_{\mathrm{lim}}@f$", color='red')
ax.plot([0, x[-1]], [T_sol[-1], T_sol[-1]], color='red')
else:
print("No such file:", T_limit_path)
if os.path.exists(lambda_limit_path):
lambda_limit = np.loadtxt(lambda_limit_path, np.float64)
ax.plot(lambda_limit[:, 0], lambda_limit[:, 1], label=r"@f$\lambda_{\mathrm{lim}}@f$", color='green')
ax.plot([0, x[-1]], [lambda_sol[-1], lambda_sol[-1]], color='green')
else:
print("No such file:", lambda_limit_path)
'''
ax.set_xlabel(r"@f$\xi@f$")
ax.set_ylabel(r"@f$u, T, \lambda@f$")
ax.legend()
plt.show()
else:
print("No such file:", filename)











